flowchart TB
A[model] --> B(assumptions)
B --> C[fit] --> D{check} -->|adequate| E(stop)
D --> |not good| B
Regression
Introduction
Regression modelsDataDistributionsFittingSelectionComparisonInterpretation
Through a simple data example
Regression models
Models and statistical modellingAssumptionsRegression ModelsDistributional RegressionExample
“www.gamlss.com”
Statistical modelling
Statistical models
“all models are wrong but some are useful”.
– George Box
Models should be parsimonious
Models should be fit for purpose and able to answer the question at hand
Statistical models have a stochastic component
All models are based on assumptions.
Assumptions
Assumptions are made to simplify things
Explicit assumptions
Implicit assumptions
it is easier to check the explicit assumptions rather the implicit ones
Model circle
Regression
- \[ X \stackrel{\textit{M}(\theta)}{\longrightarrow} Y \]
- \(y\): targer, the y or the dependent variable
- \(X\): explanatory, features, x’s or independent variables or terms
Linear Model
- standard way
\[ \begin{equation} y_i= b_0 + b_1 x_{1i} + b_2 x_{2i}, \ldots, b_p x_{pi}+ \epsilon_i \end{equation} \qquad(1)\]
Linear Model
- different way
\[ \begin{eqnarray} y_i & \stackrel{\small{ind}}{\sim } & {N}(\mu_i, \sigma) \nonumber \\ \mu_i &=& b_0 + b_1 x_{1i} + b_2 x_{2i}, \ldots, b_p x_{pi} \end{eqnarray} \qquad(2)\]
Additive Models
\[ \begin{eqnarray} y_i & \stackrel{\small{ind}}{\sim } & {N}(\mu_i, \sigma) \nonumber \\ \mu_i &=& b_0 + s_1(x_{1i}) + s_2(x_{2i}), \ldots, s_p(x_{pi}) \end{eqnarray} \qquad(3)\]
Machine Learning Models
\[\begin{eqnarray} y_i & \stackrel{\small{ind}}{\sim }& {N}(\mu_i, \sigma) \nonumber \\ \mu_i &=& ML(x_{1i},x_{2i}, \ldots, x_{pi}) \end{eqnarray} \qquad(4)\]
Generalised Linear Models
\[\begin{eqnarray} y_i & \stackrel{\small{ind}}{\sim }& {E}(\mu_i, \phi) \nonumber \\ g(\mu_i) &=& b_0 + b_1 x_{1i} + b_2 x_{2i}, \ldots, b_p x_{pi} \end{eqnarray} \qquad(5)\]
\({E}(\mu_i, \phi)\) :
Exponentialfamily\(g(\mu_i)\) : the
linkfunction
Distributional regression
Distributional regression
\[ X \stackrel{\textit{M}(\boldsymbol{\theta})}{\longrightarrow} D\left(Y|\boldsymbol{\theta}(\textbf{X})\right) \]
All parameters \(\boldsymbol{\theta}\) could functions of the explanatory variables \(\boldsymbol{\theta}(\textbf{X})\).
\(D\left(Y|\boldsymbol{\theta}(\textbf{X})\right)\) can be any \(k\) parameter distribution
Generalised Additive models for Location Scale and Shape
\[\begin{eqnarray} y_i & \stackrel{\small{ind}}{\sim }& {D}( \theta_{1i}, \ldots, \theta_{ki}) \nonumber \\ g(\theta_{1i}) &=& b_{10} + s_1({x}_{1i}) + \ldots, s_p({x}_{pi}) \nonumber\\ \ldots &=& \ldots \nonumber\\ g({\theta}_{ki}) &=& b_0 + s_1({x}_{1i}) + \ldots, s_p({x}_{pi}) \end{eqnarray} \qquad(6)\]
GAMLSS + ML
\[\begin{eqnarray} y_i & \stackrel{\small{ind}}{\sim }& {D}( \theta_{1i}, \ldots, \theta_{ki}) \nonumber \\ g({\theta}_{1i}) &=& {ML}_1({x}_{1i},{x}_{2i}, \ldots, {x}_{pi}) \nonumber \\ \ldots &=& \ldots \nonumber\\ g({\theta}_{ki}) &=& {ML}_1({x}_{1i},{x}_{2i}, \ldots, {x}_{pi}) \end{eqnarray} \qquad(7)\]
Example
Figure 1 Abdominal circumference against gestation age.
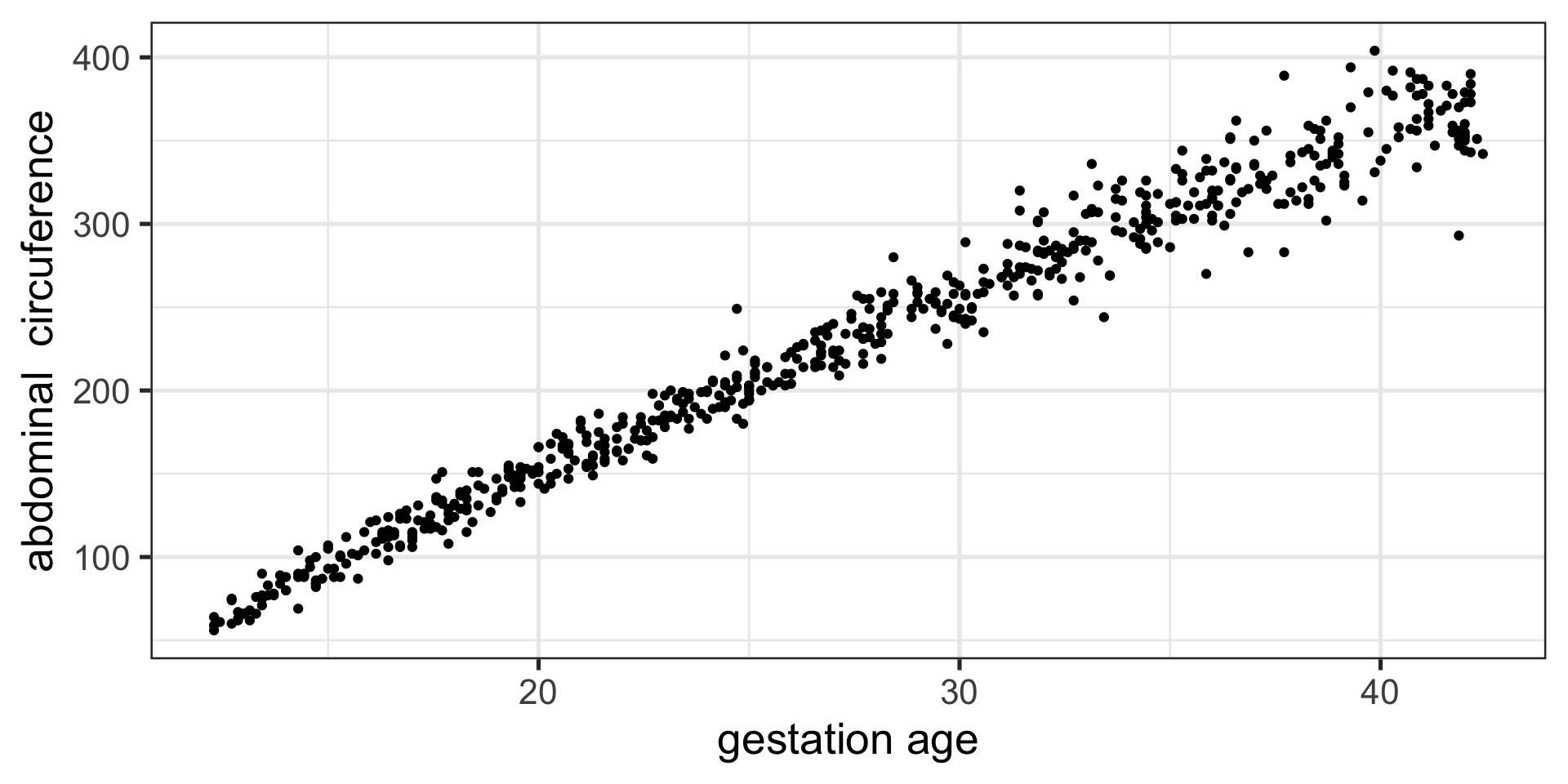
Figure 1: The abdom data.
Fitting Models
library(ggplot2)
library(gamlss.ggplots)
library(gamlss.add)
# Linear
lm1 <- gamlss(y~x, data=abdom, trace=FALSE)
# additive smooth
am1 <- gamlss(y~pb(x), data=abdom,trace=FALSE)# smooth
# neural network
set.seed(123)
nn1 <- gamlss(y~nn(~x), size=5, data=abdom, trace=FALSE)# neural
# regression three
rt1 <- gamlss(y~tr(~x), data=abdom, trace=FALSE)# three
GAIC(lm1, am1, nn1, rt1)Fitting Models
df AIC
am1 6.508274 4948.869
nn1 12.000000 4965.171
lm1 3.000000 5008.453
rt1 14.000000 5305.390Linear Model
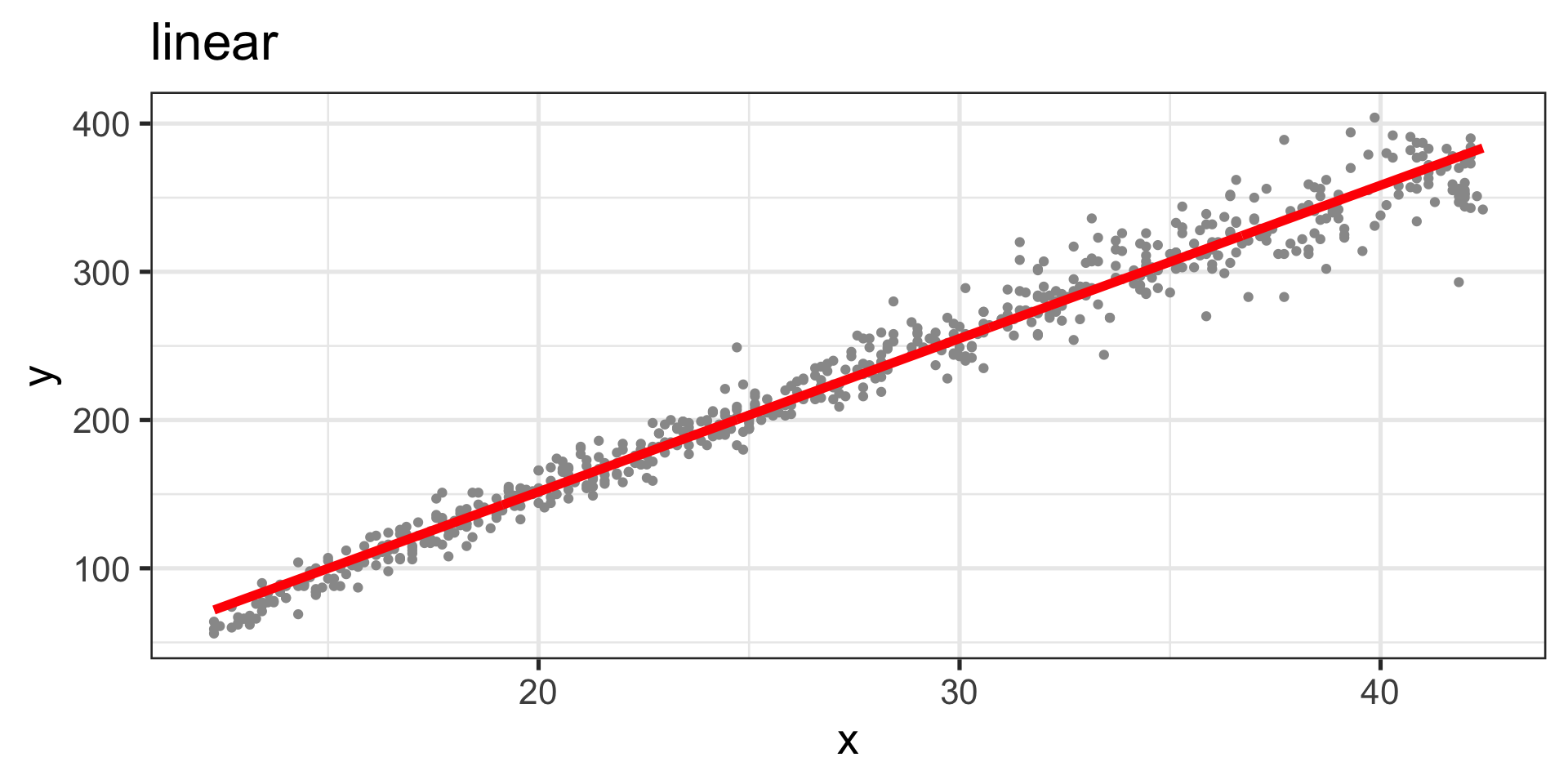
Figure 2: Fitted values, linear curve
Additive Smooth Model
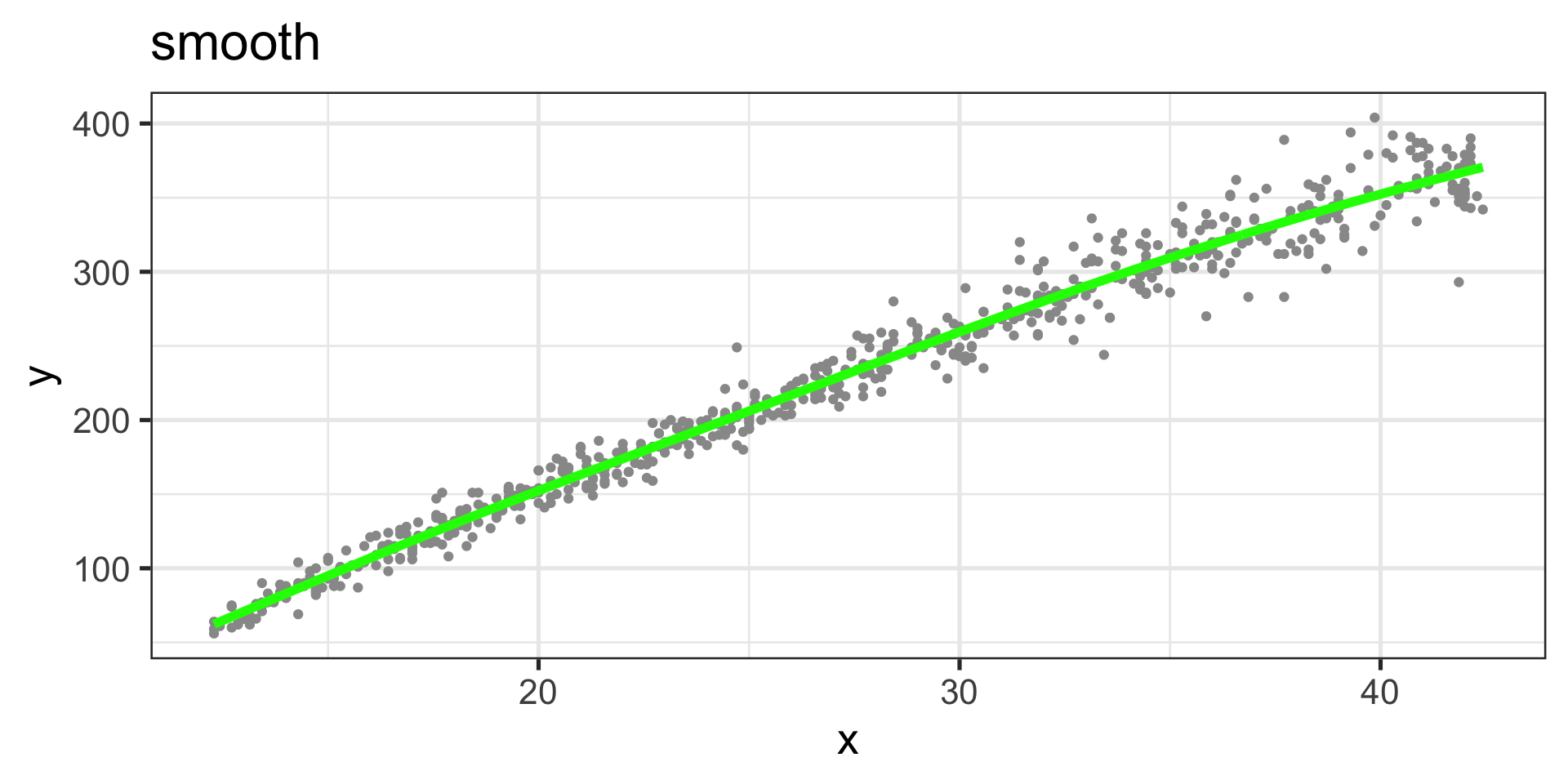
Figure 3: Fitted values, smooth curve
Neural network
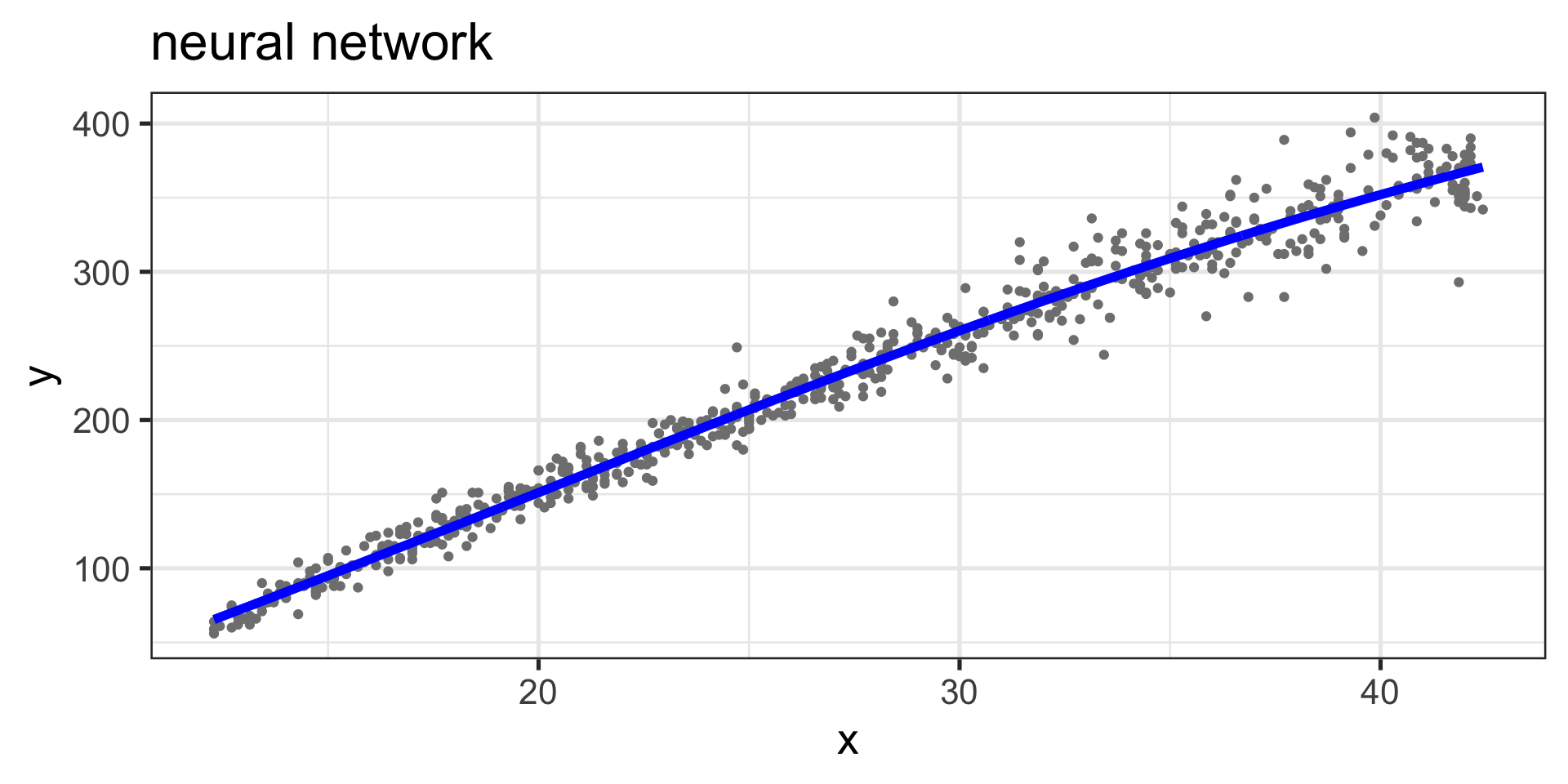
Figure 4: Fitted values, neural network curve
Regression Tree
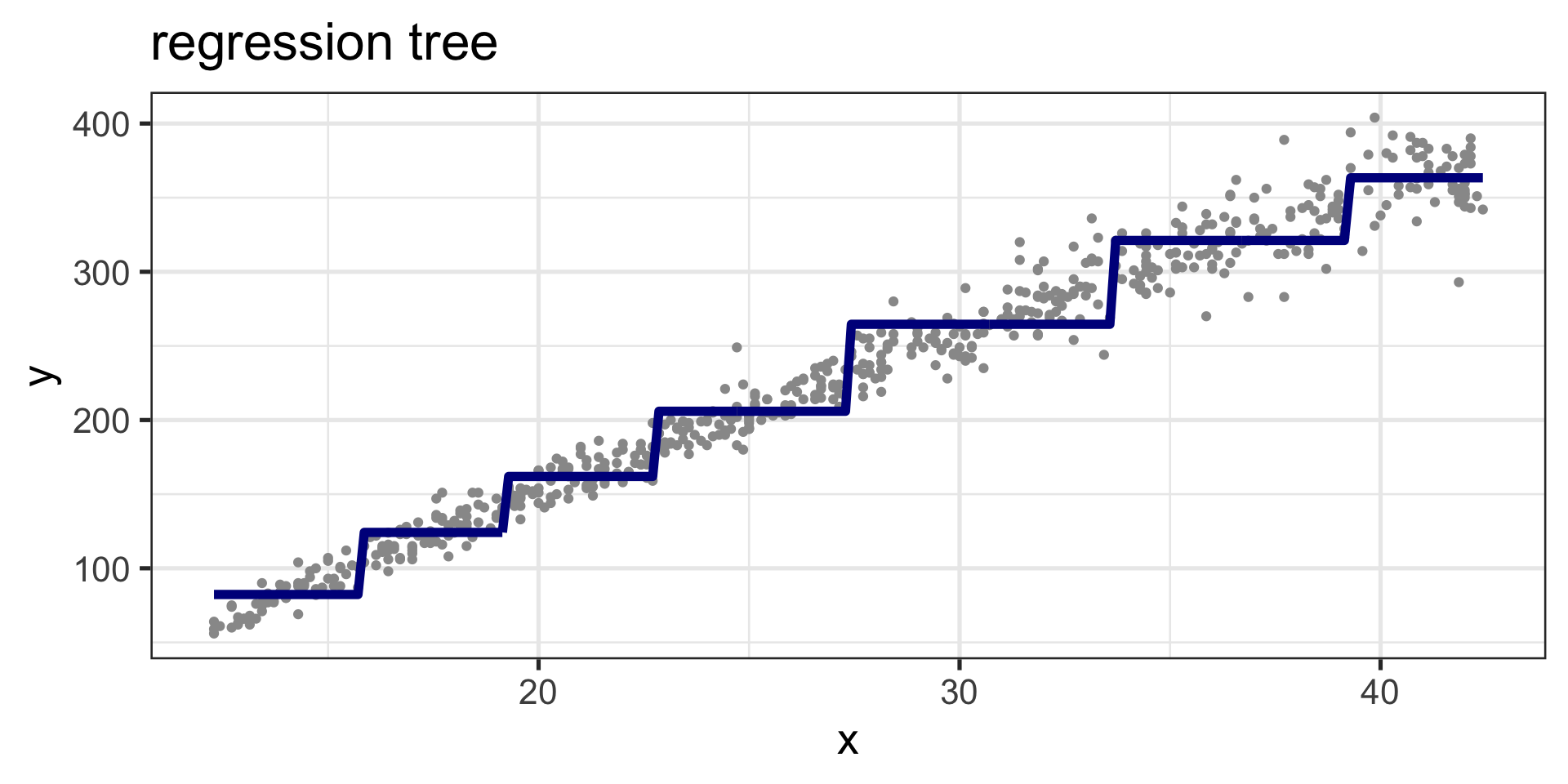
Figure 5: Fitted values, regression tree curve
Diagnostics: QQ plot
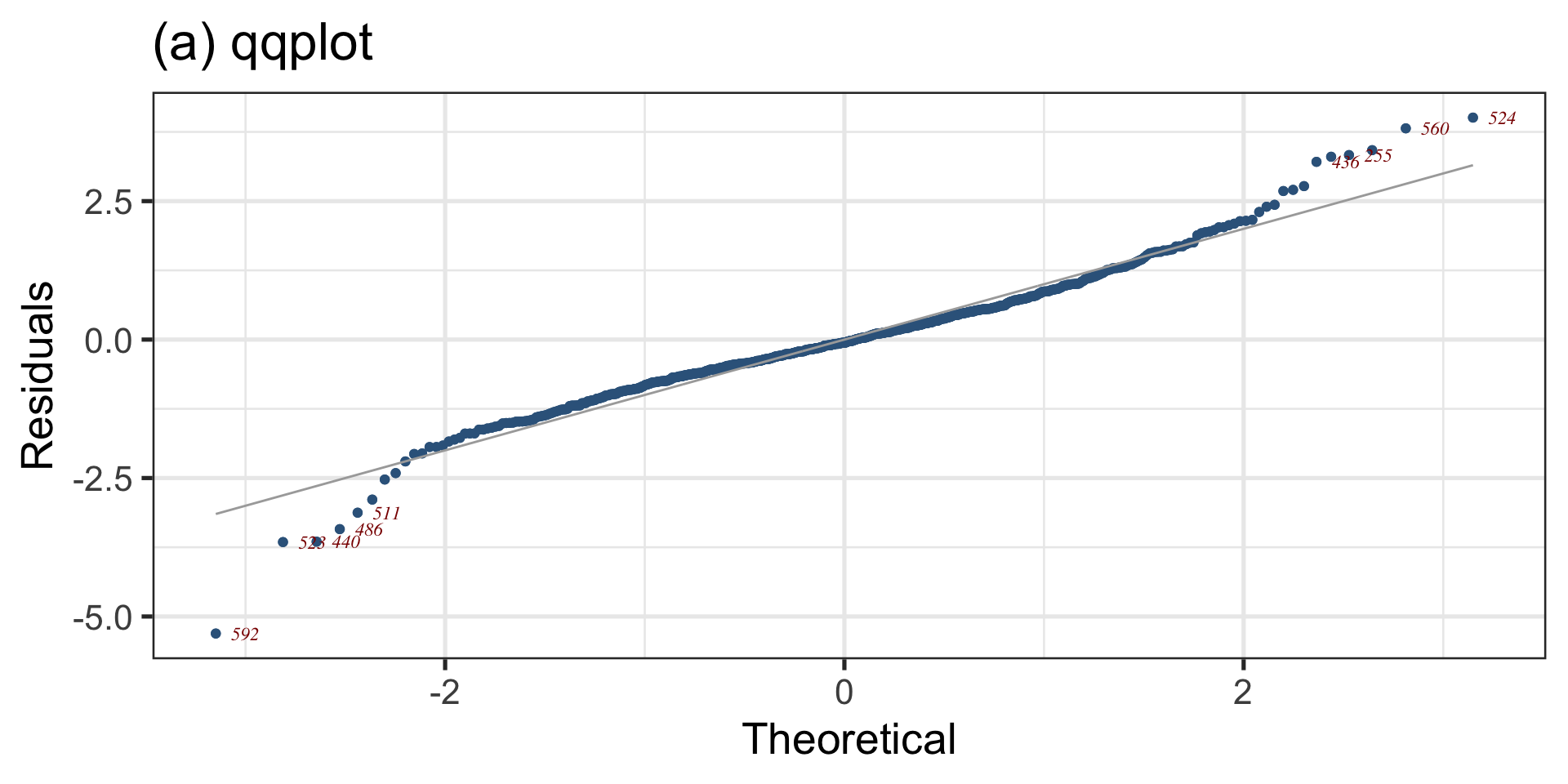
Figure 6: QQ-plot of the fitted am1 model
Diagnostics: Bucket plot

Figure 7: QQ-plot of the fitted am1 model
Refit
QQ plot, Logistic
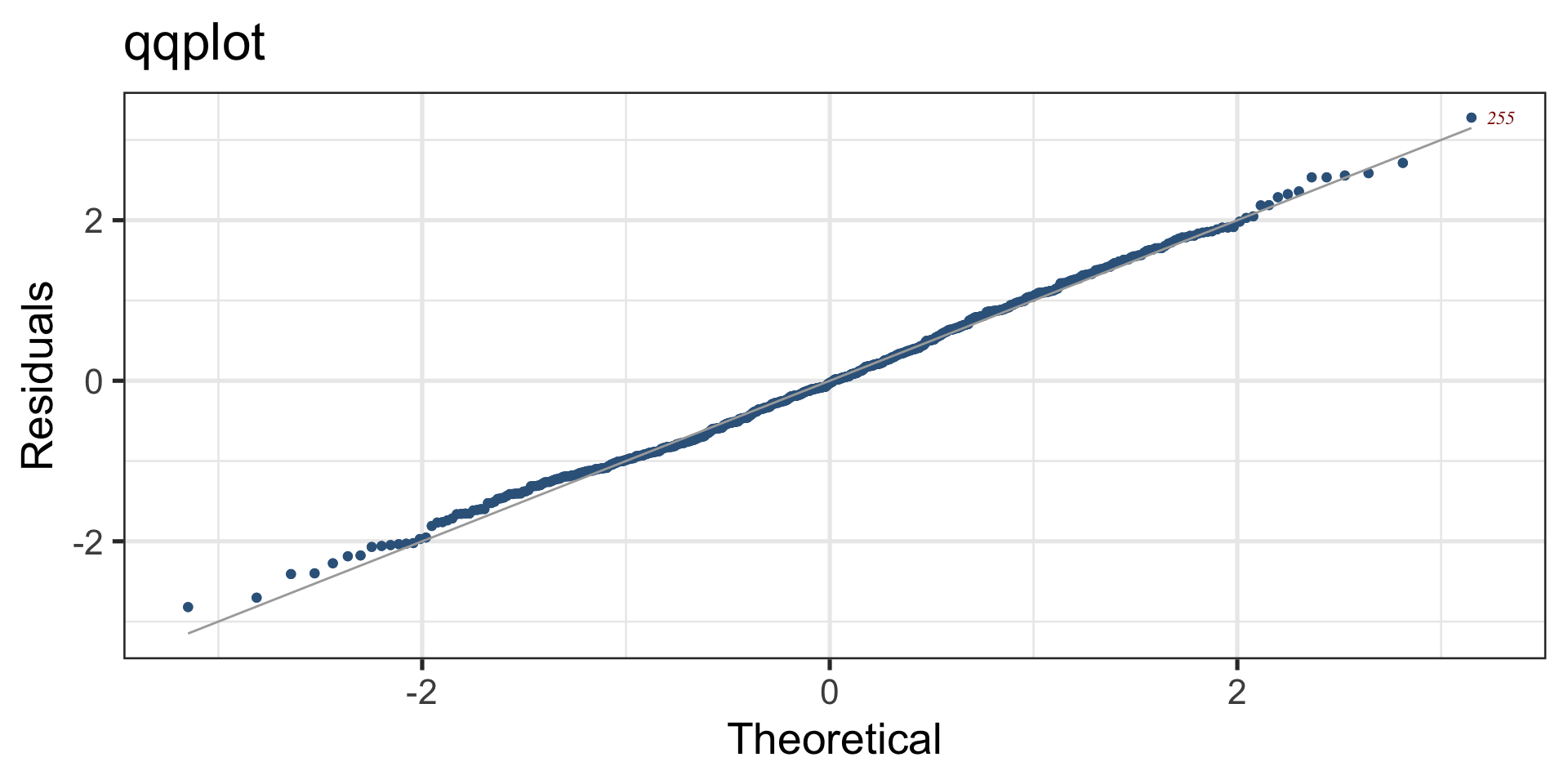
Figure 8: QQ-plot of the fitted am1 model
Bucket plot, Logistic
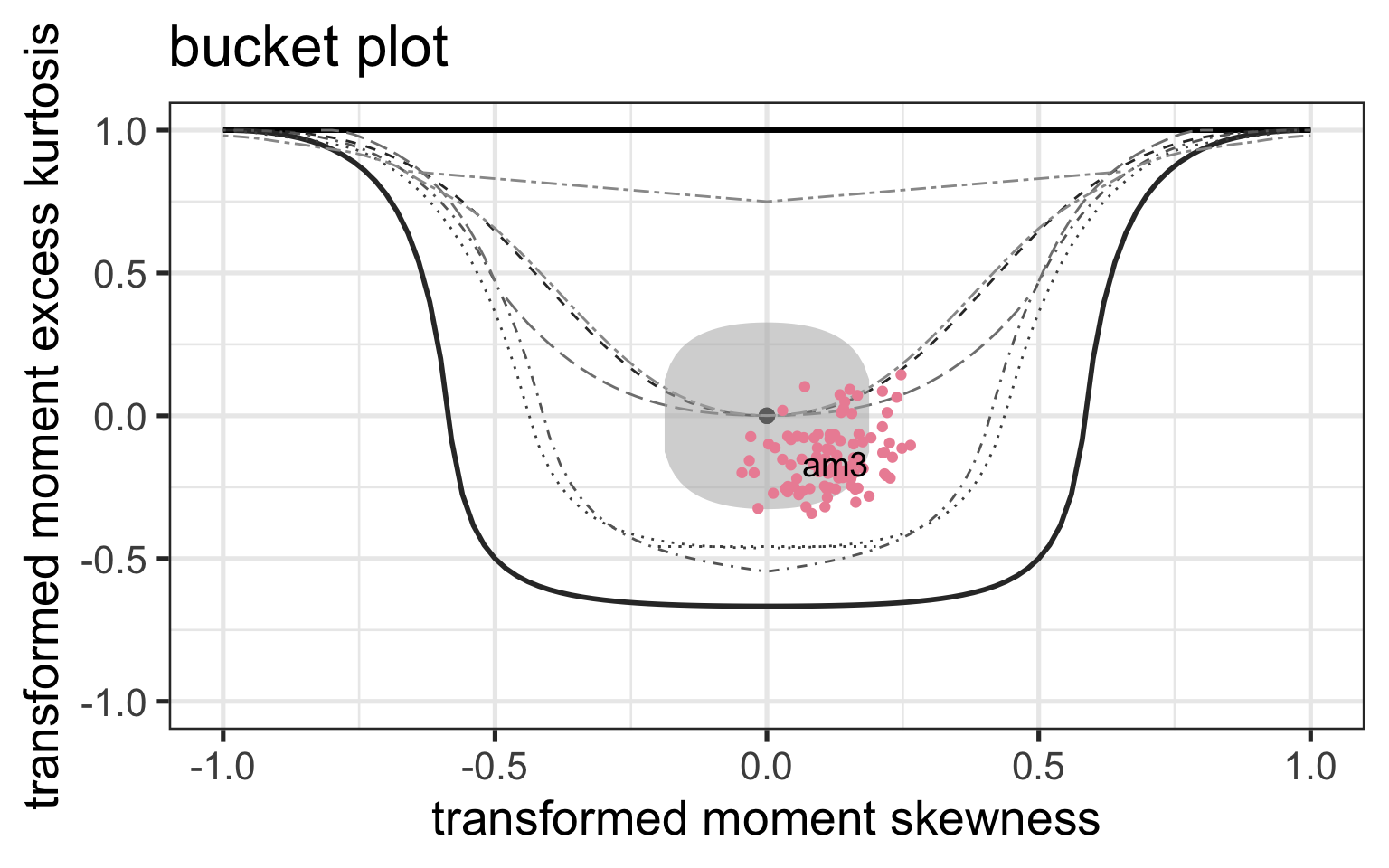
Figure 9: QQ-plot of the fitted am1 model
Fitted Centiles
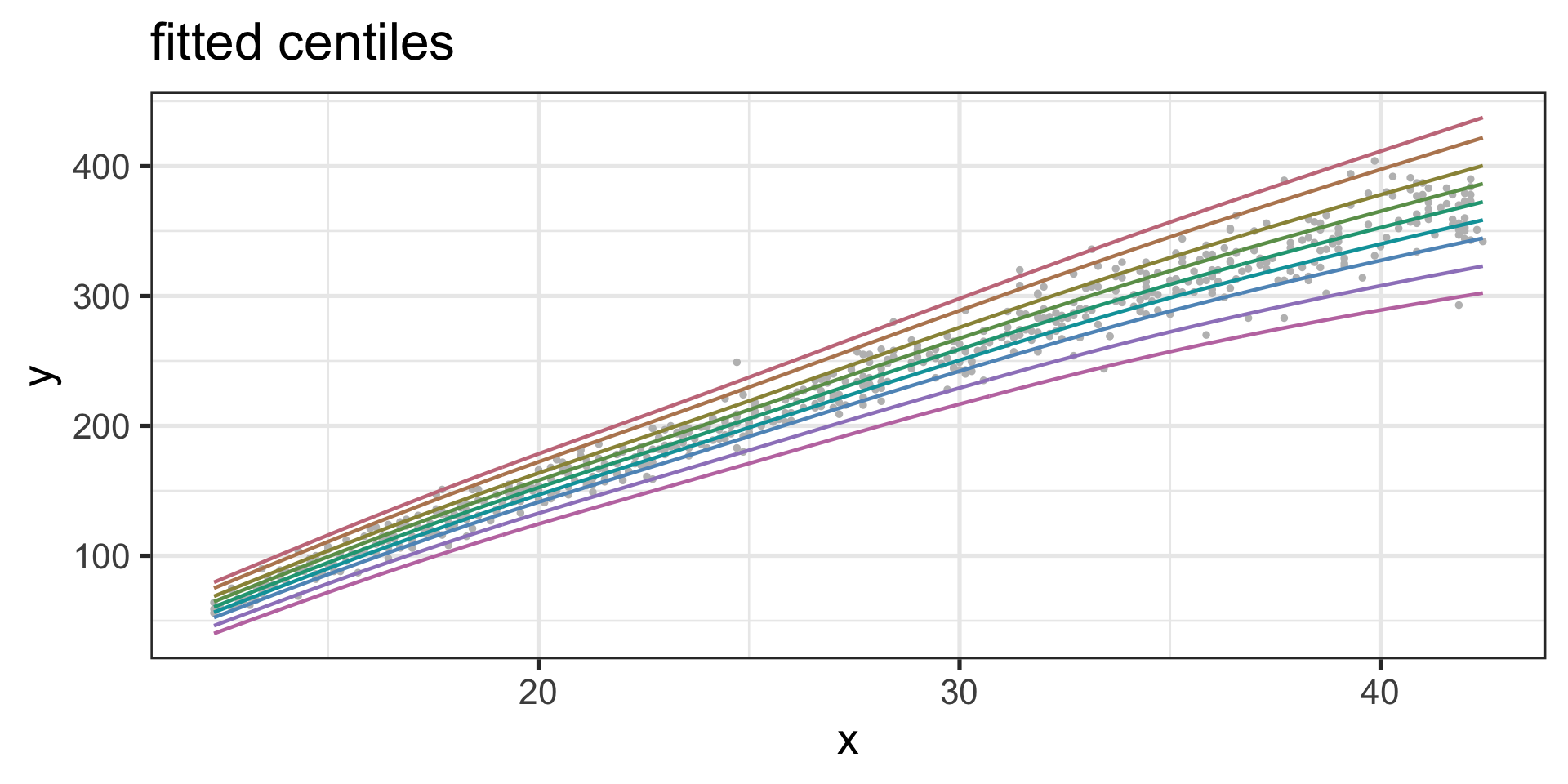
Figure 10: Centile-plot of the fitted am1 model
Fitted Distributions
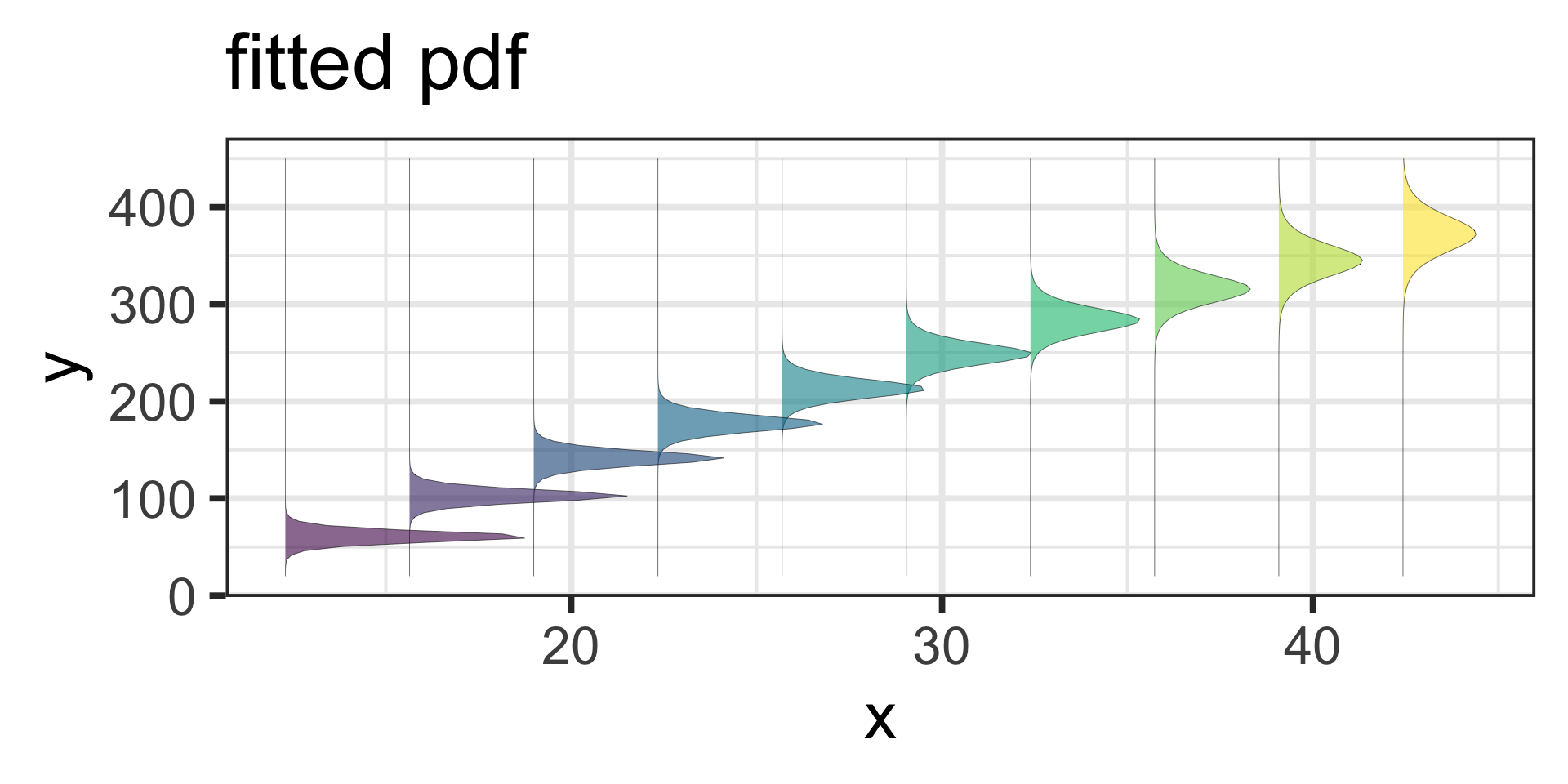
Figure 11: pdf-plot of the fitted am3 model
Summary
The additive smooth model is the best parsimonious model
A kurtotic distribution is adequate for the data
No simple Machine Learning method will do because there is kurtosis and we are interested in centiles
quantile regressioncould be used here but in general it is more difficult to check the implicit assumptions made
Tip
Implicit assumptions are more difficult to check
end


 The Books
The Books

www.gamlss.com