Fitting
Mikis Stasinopoulos
Bob Rigby
Fernanda De Bastiani
Introduction
the basic GAMLSS algorithmdifferent statistical approaches for fitting a GAMLSS modeldifferent machine learning techniques for fitting a GAMLSS model.
Machine Learning Models
properties
are the standard errors available?
do the x’s need standardization.
about the algorithm
- stability - speed - convergencenonlinear terms
interactions
dataset type
is the selection of x’s automatic?
interpretation
Linear Models
Linear Models
******************************************************************
Family: c("BCTo", "Box-Cox-t-orig.")
Call: gamlss(formula = rent ~ area + poly(yearc, 2) + location +
bath + kitchen + cheating, sigma.formula = ~area +
yearc + location + bath + kitchen + cheating, family = BCTo,
data = da, trace = FALSE)
Fitting method: RS()
------------------------------------------------------------------
Mu link function: log
Mu Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 5.0138582 0.0277852 180.451 < 2e-16 ***
area 0.0106311 0.0002331 45.613 < 2e-16 ***
poly(yearc, 2)1 5.0382576 0.3360044 14.995 < 2e-16 ***
poly(yearc, 2)2 3.3475762 0.2737237 12.230 < 2e-16 ***
location2 0.0875504 0.0102485 8.543 < 2e-16 ***
location3 0.1967006 0.0385032 5.109 3.44e-07 ***
bath1 0.0415393 0.0219353 1.894 0.0584 .
kitchen1 0.1120385 0.0243199 4.607 4.25e-06 ***
cheating1 0.3297044 0.0243422 13.545 < 2e-16 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
------------------------------------------------------------------
Sigma link function: log
Sigma Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 10.4022601 0.7528378 13.817 < 2e-16 ***
area 0.0012116 0.0006135 1.975 0.0484 *
yearc -0.0059332 0.0003864 -15.356 < 2e-16 ***
location2 0.0595118 0.0254048 2.343 0.0192 *
location3 0.2191754 0.0930995 2.354 0.0186 *
bath1 0.0058254 0.0087229 0.668 0.5043
kitchen1 0.0400246 0.0882200 0.454 0.6501
cheating1 -0.2453139 0.0487224 -5.035 5.06e-07 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
------------------------------------------------------------------
Nu link function: identity
Nu Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 0.65631 0.05598 11.72 <2e-16 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
------------------------------------------------------------------
Tau link function: log
Tau Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 3.2918 0.4411 7.463 1.09e-13 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
------------------------------------------------------------------
No. of observations in the fit: 3082
Degrees of Freedom for the fit: 19
Residual Deg. of Freedom: 3063
at cycle: 10
Global Deviance: 38254.51
AIC: 38292.51
SBC: 38407.14
******************************************************************Additive Models
Additive Models
df AIC
mAM 29.78058 38198.79
mLM 19.00000 38292.51Additive Models (continue)
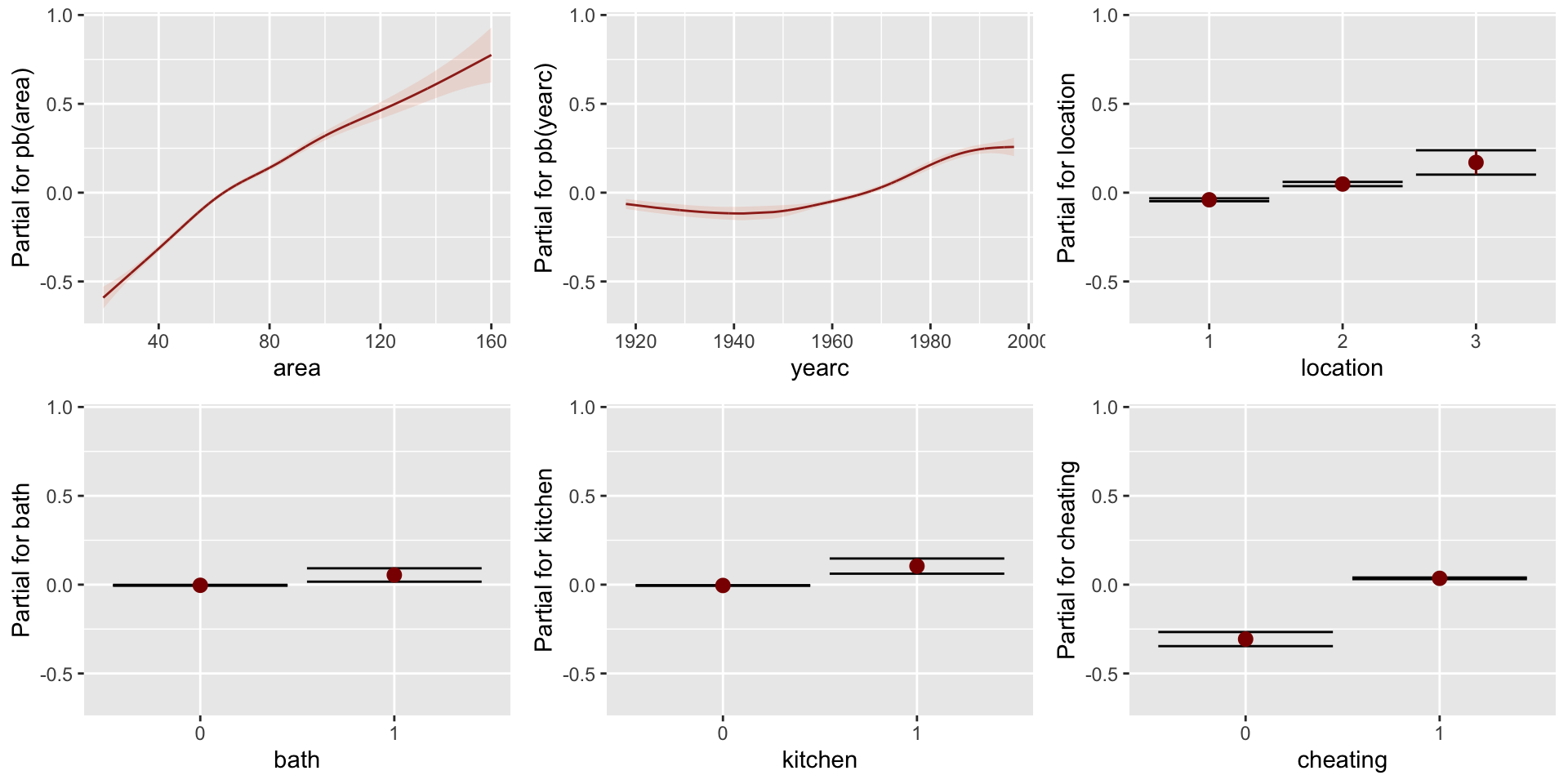
Figure 1: pdf-plot of the fitted am1 mu model
Additive Models (continue)
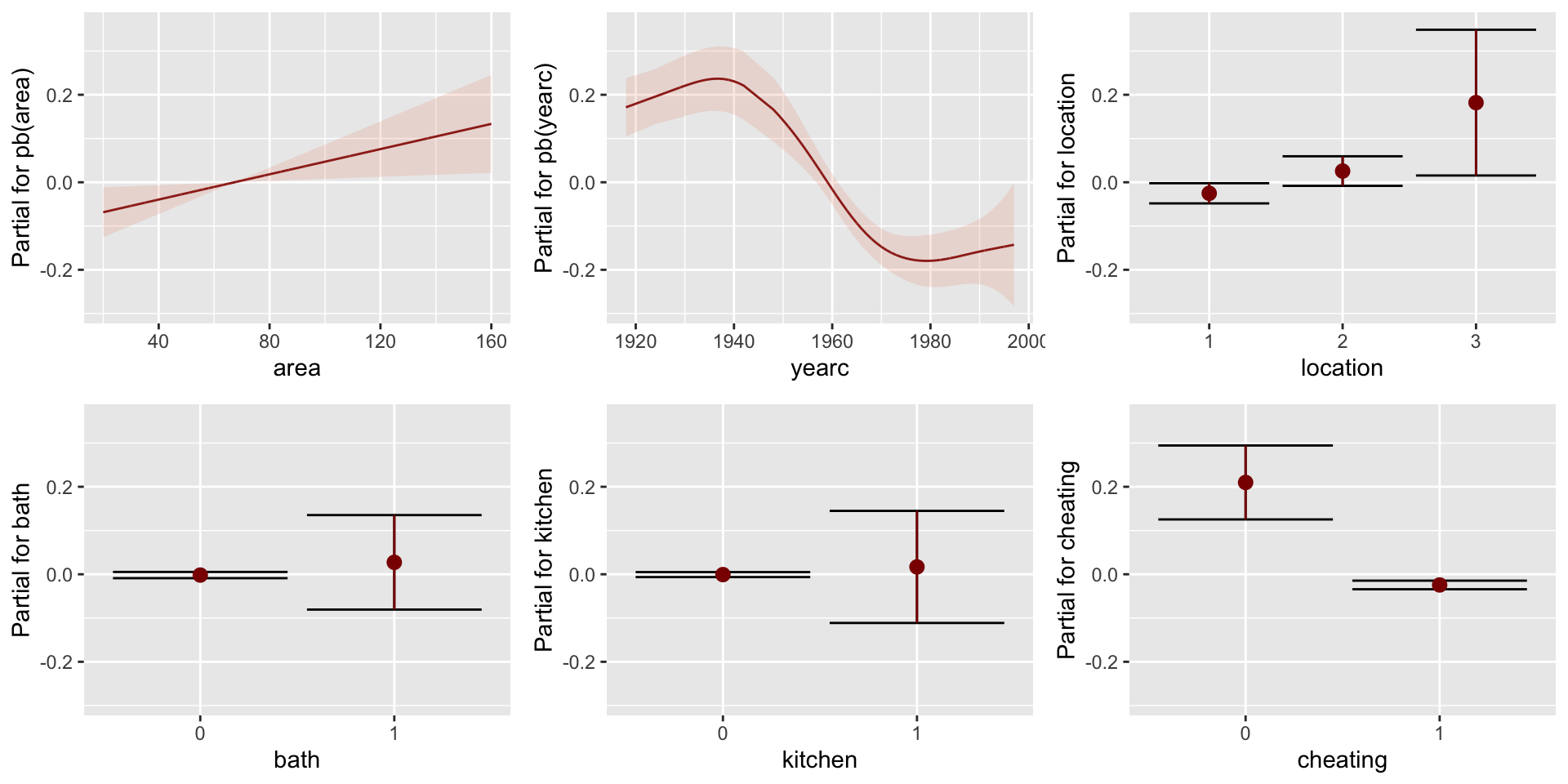
Figure 2: pdf-plot of the fitted am1 sigma model
Regression Trees
Regression Trees
df AIC
mAM 29.78058 38198.79
mLM 19.00000 38292.51
mRT 22.00000 38711.66Regression Trees (continue)
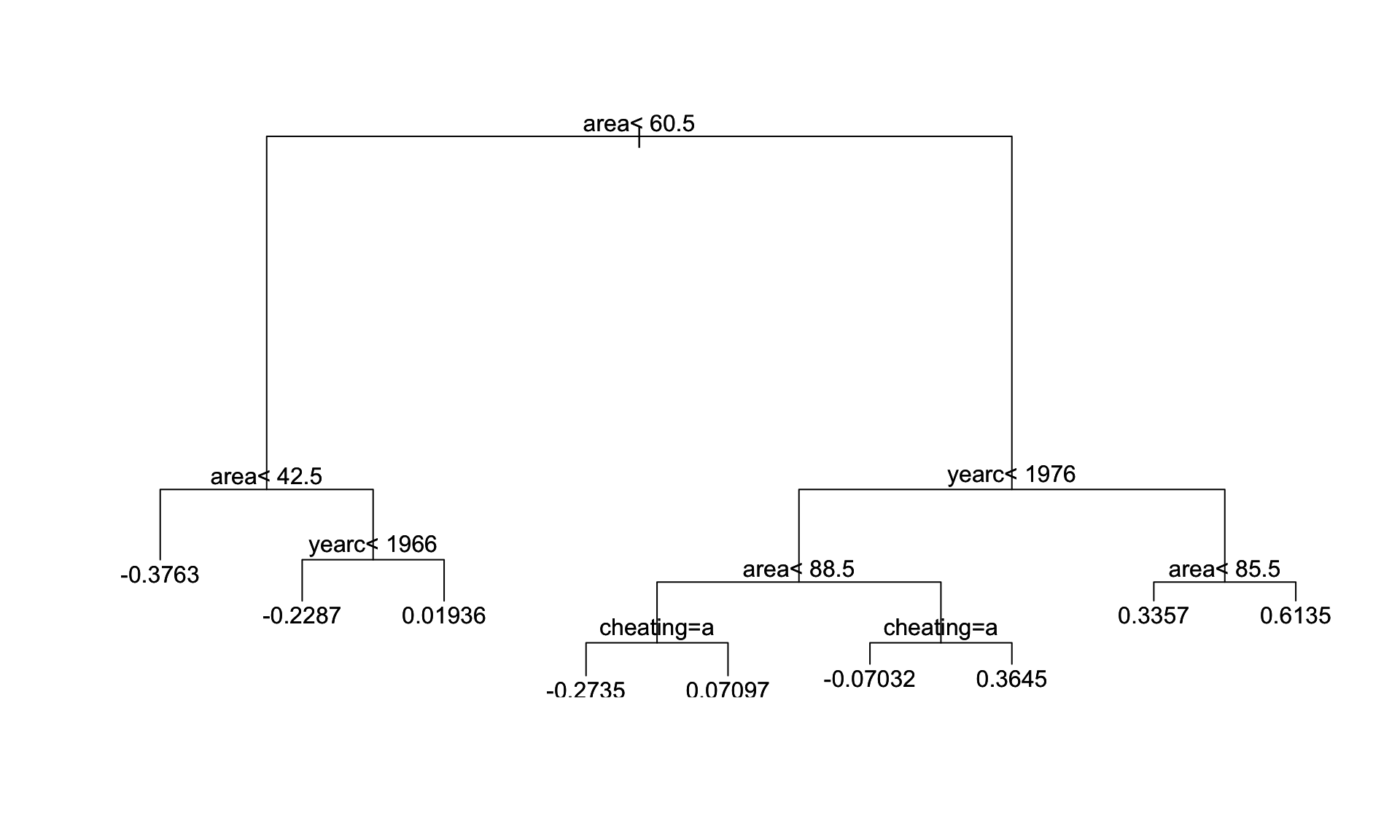
Figure 3: The fitted tree for mu model
Regression Trees (continue)
Figure 4: The fitted tree for mu model
Neural Networks
Neural Networks
GAMLSS-RS iteration 1: Global Deviance = 38089.89
GAMLSS-RS iteration 2: Global Deviance = 38054.95
GAMLSS-RS iteration 3: Global Deviance = 38054.1
GAMLSS-RS iteration 4: Global Deviance = 38053.9
GAMLSS-RS iteration 5: Global Deviance = 38053.84
GAMLSS-RS iteration 6: Global Deviance = 38053.79
GAMLSS-RS iteration 7: Global Deviance = 38053.78
GAMLSS-RS iteration 8: Global Deviance = 38053.78
GAMLSS-RS iteration 9: Global Deviance = 38053.78 df AIC
mAM 29.78058 38198.79
mNN 78.00000 38209.78
mLM 19.00000 38292.51
mRT 22.00000 38711.66Neural Networks (continue)
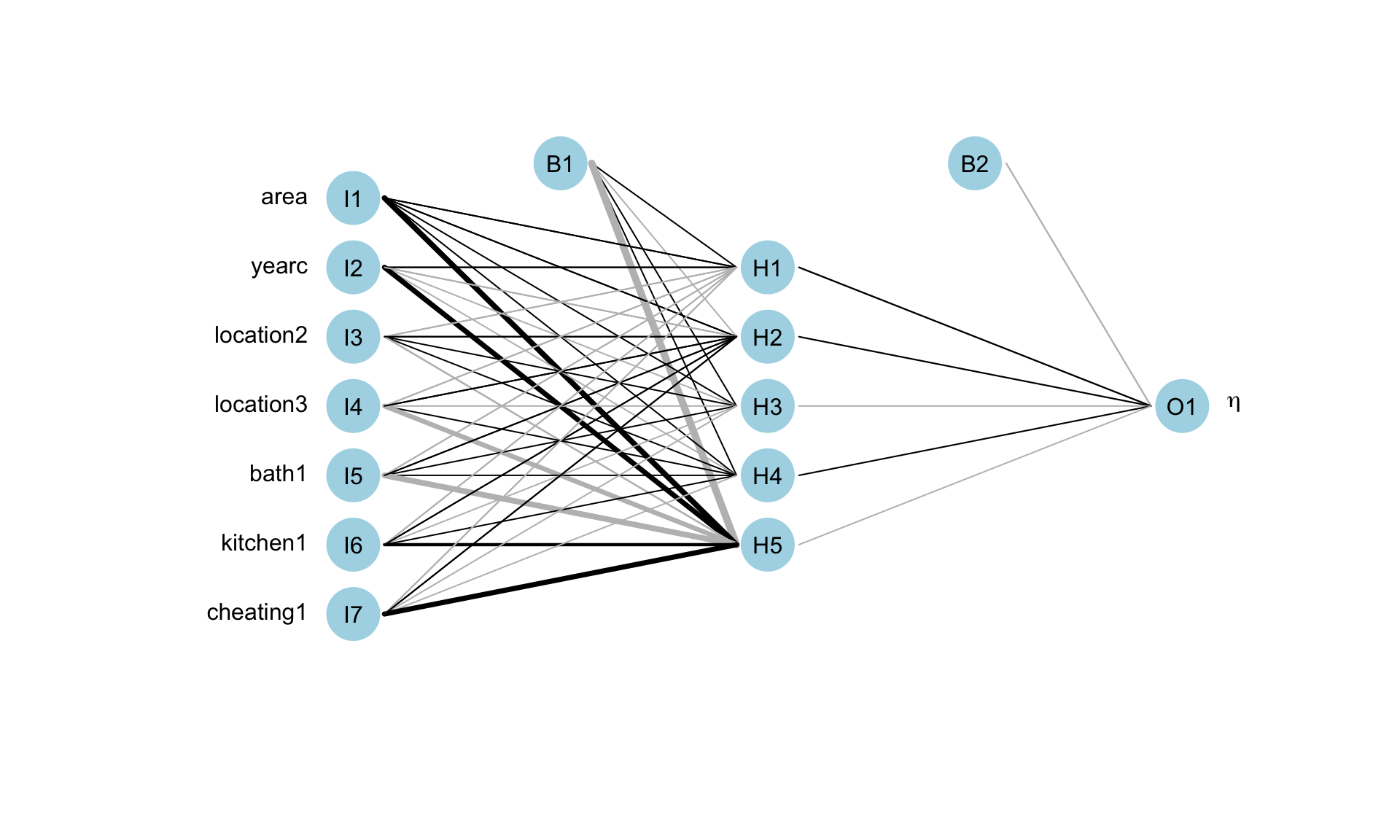
Figure 5: The fitted neural network model fot mu.
Neural Networks
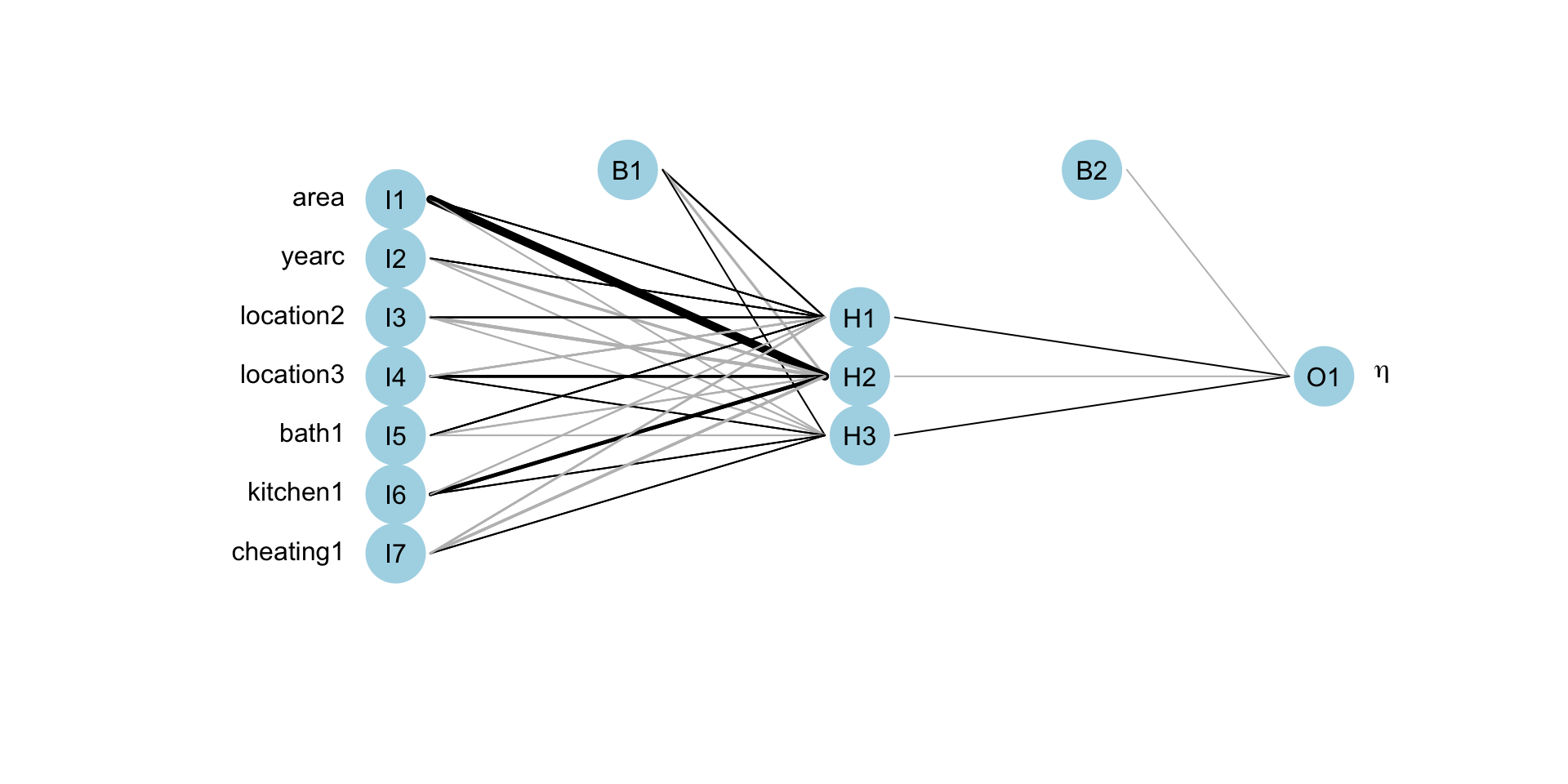
Figure 6: The fitted neural network model fot sigma.
LASSO
library(gamlss.lasso)
source("~/Dropbox/github/gamlss-ggplots/R/data_stand.R")
da10 <- data_scale(da, response=rent)
da1 <- data_form2df(da10, response=rent, type="main.effect",
nonlinear="TRUE", arg=3)
mLASSO <- gamlss(rent~gnet(x.vars=names(da1)[-c(1)],
method = "IC", ICpen="BIC"),
~gnet(x.vars=names(da1)[-c(1)],
method = "IC", ICpen="BIC"),
data=da1, family=BCTo, bf.cyc=1,
c.crit=0.1, trace=FALSE)
AIC(mLM, mAM, mRT, mNN, mLASSO)LASSO
df AIC
mAM 29.78058 38198.79
mNN 78.00000 38209.78
mLM 19.00000 38292.51
mLASSO 22.00000 38309.11
mRT 22.00000 38711.66LASSO (continue)
LASSO (continue)
area1 area2 area3 yearc1 yearc2 yearc3
13.72498782 -1.85175963 0.09162263 4.97838839 3.71690657 -0.18975745
location2 location3 cheating1
0.01331635 0.05631020 0.27463624 area1 area2 area3 yearc1 yearc3 location2 location3
2.2850872 -0.5173168 1.6240863 -7.7665983 3.2776033 0.0673355 0.3178812
kitchen1 cheating1
0.1209266 -0.1497227 Principal Component Regression
library(gamlss.foreach)
registerDoParallel(cores = 10)
source("~/Dropbox/github/gamlss-ggplots/R/data_stand.R")
source("~/Dropbox/GAMLSS-development/PCR/GAMLSS-pc.R")
X = formula2X(rent~area+yearc+location+bath+kitchen+
cheating, data=da)
mPCR <- gamlss(rent~pc(x=X),~pc(x=X),
data=da1, family=BCTo, bf.cyc=1, trace=TRUE)Principal Component Regression
GAMLSS-RS iteration 1: Global Deviance = 38458.91
GAMLSS-RS iteration 2: Global Deviance = 38404.13
GAMLSS-RS iteration 3: Global Deviance = 38405.09
GAMLSS-RS iteration 4: Global Deviance = 38404.56
GAMLSS-RS iteration 5: Global Deviance = 38404.43
GAMLSS-RS iteration 6: Global Deviance = 38404.38
GAMLSS-RS iteration 7: Global Deviance = 38404.37
GAMLSS-RS iteration 8: Global Deviance = 38404.36
GAMLSS-RS iteration 9: Global Deviance = 38404.36
GAMLSS-RS iteration 10: Global Deviance = 38404.36 df AIC
mAM 29.78058 38198.79
mNN 78.00000 38209.78
mLM 19.00000 38292.51
mLASSO 22.00000 38309.11
mPCR 15.00000 38434.36
mRT 22.00000 38711.66summary
| ML Models | coef. s.e. | stand. of x’s | algo. stab., speed, conv. | non-linear terms | inter- actions | data type | auto sele-ction | interpre- tation |
|---|---|---|---|---|---|---|---|---|
| linear | yes | no | yes, fast, v.good | poly | declare | \(n>r\) | no | v. easy |
| additive | no | no | yes, slow, good | smooth | declare | \(n>r\) | no | easy |
| RT | no | no | no, slow, bad | trees | auto | \(n>r\)?? | yes | easy |
summary (continue)
| ML Models | coef. s.e. | stand. of x’s | algo. stab., speed, conv. | non-linear terms | inter- actions | data type | auto sele-ction | interpre- tation |
|---|---|---|---|---|---|---|---|---|
| NN | no | 0 to 1 | no, \(\,\,\) ok, \(\,\,\) ok | auto | auto | both? | yes | v. hard |
| PCR | yes | yes | yes, fast, good | poly | declare | both | auto | hard |
| LASSO | no | yes | yes, fast, good | poly | declare | both | auto | easy |
summary (continue)
| ML Models | coef. s.e. | stand. of x’s | algo. stab., speed, conv. | non-linear terms | inter- actions | data type | auto sele-ction | interpre- tation |
|---|---|---|---|---|---|---|---|---|
| Boost | no | no | yes, fast, good | smooth trees | declare | \(n<<r\) | yes | easy |
| MCMC | yes | no | good, ok, \(\,\,\) ok | smooth | declare | \(n>r\) | no | easy |
diagram
flowchart TB A[Data] --> B(n greater than r) A --> C( n less than r) B --> D[LM,GLM, \n GAM, NN ] C --> E[RidgeR, LASSO, Elastic Net, \n PCR, RF, \n Boosting, NN]
end


 The Books
The Books

www.gamlss.com