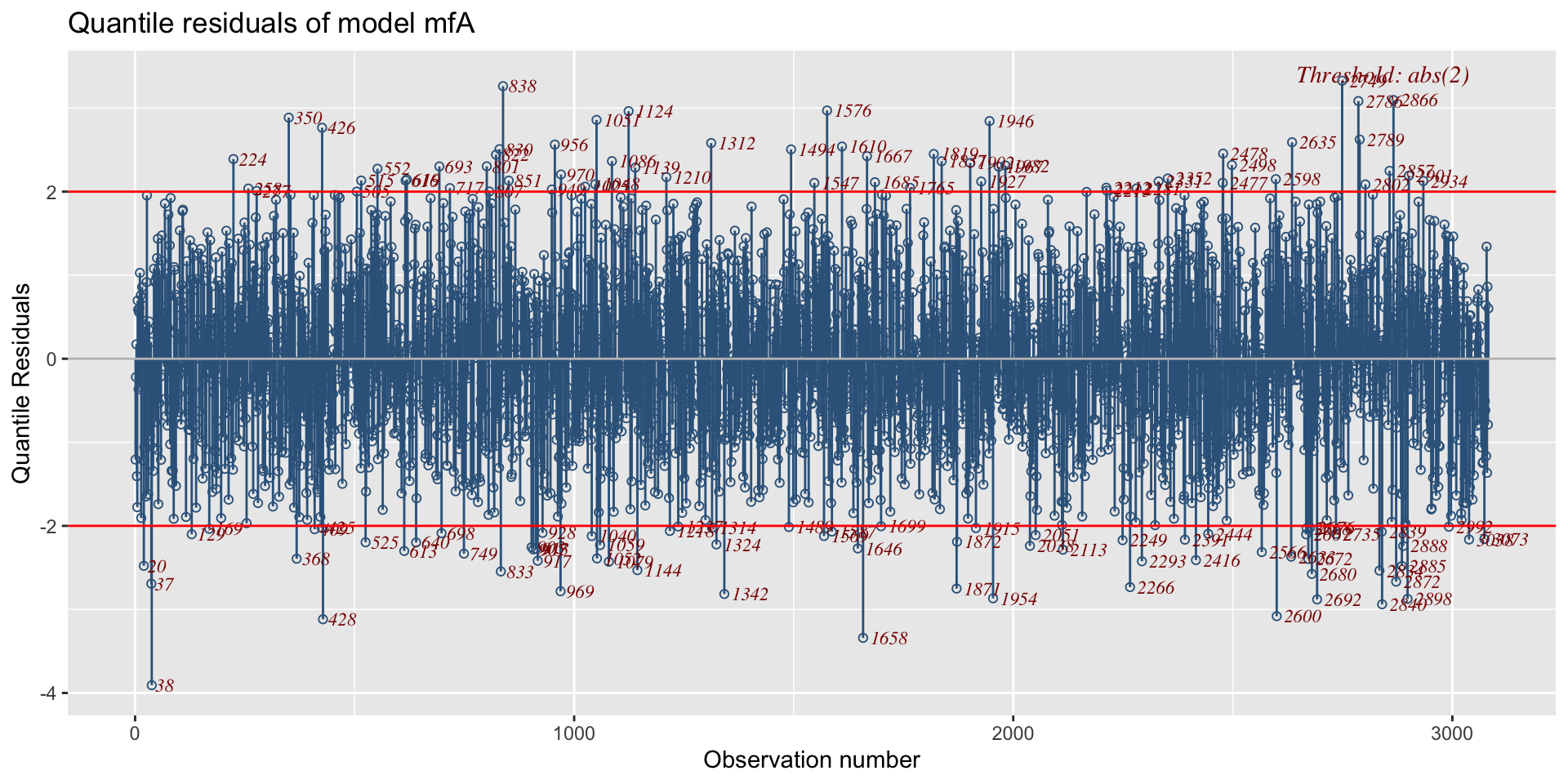
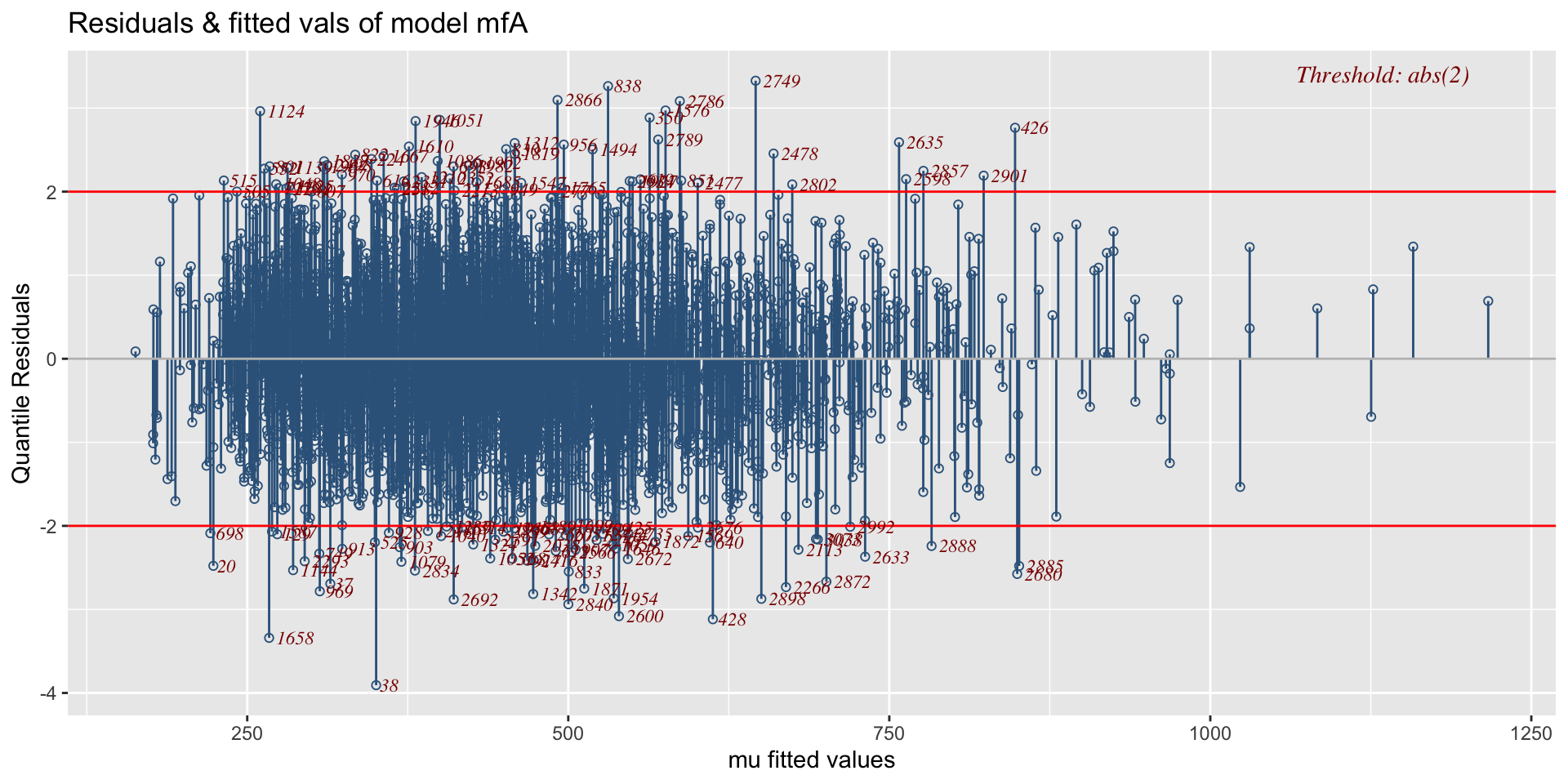
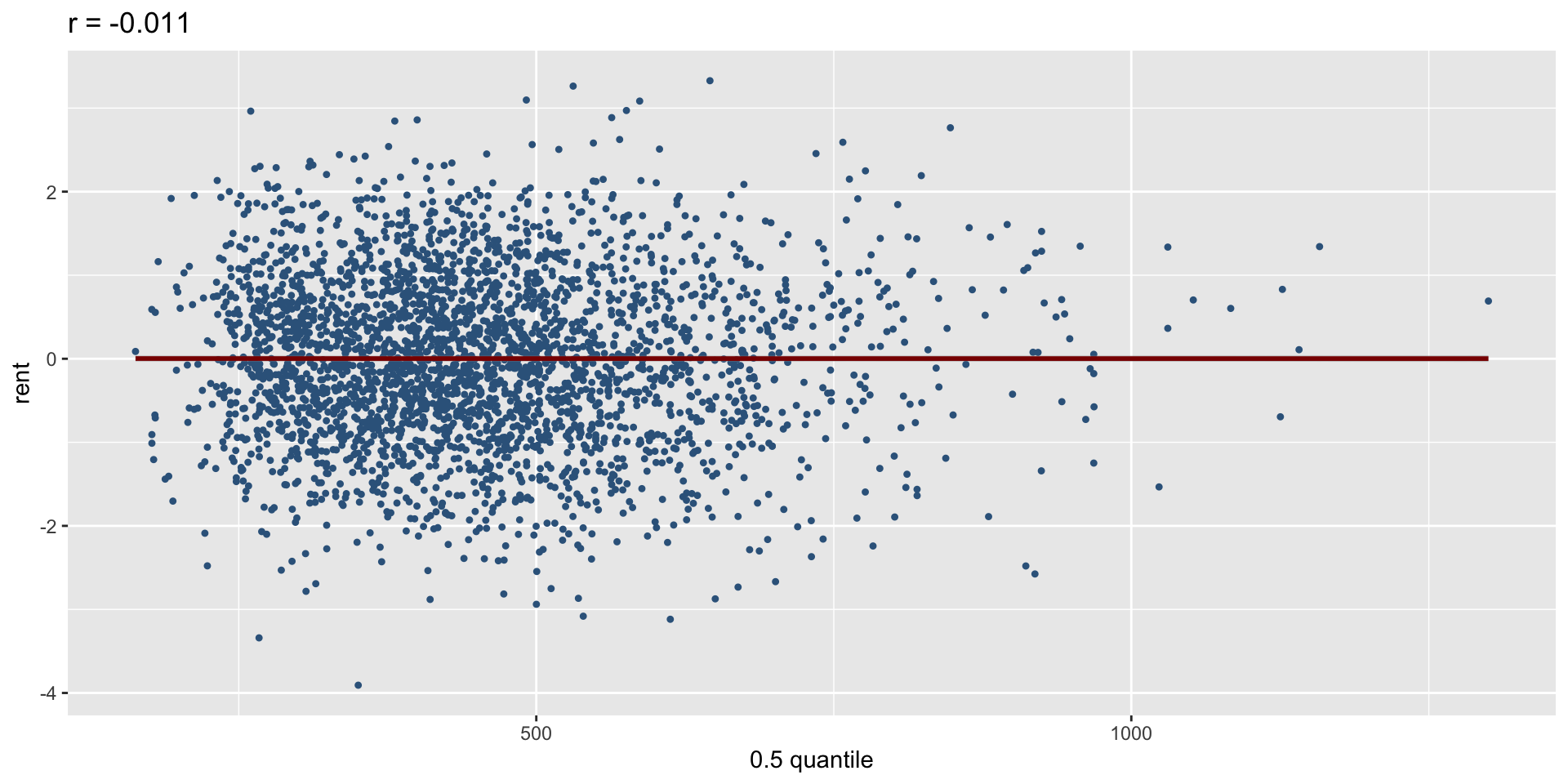
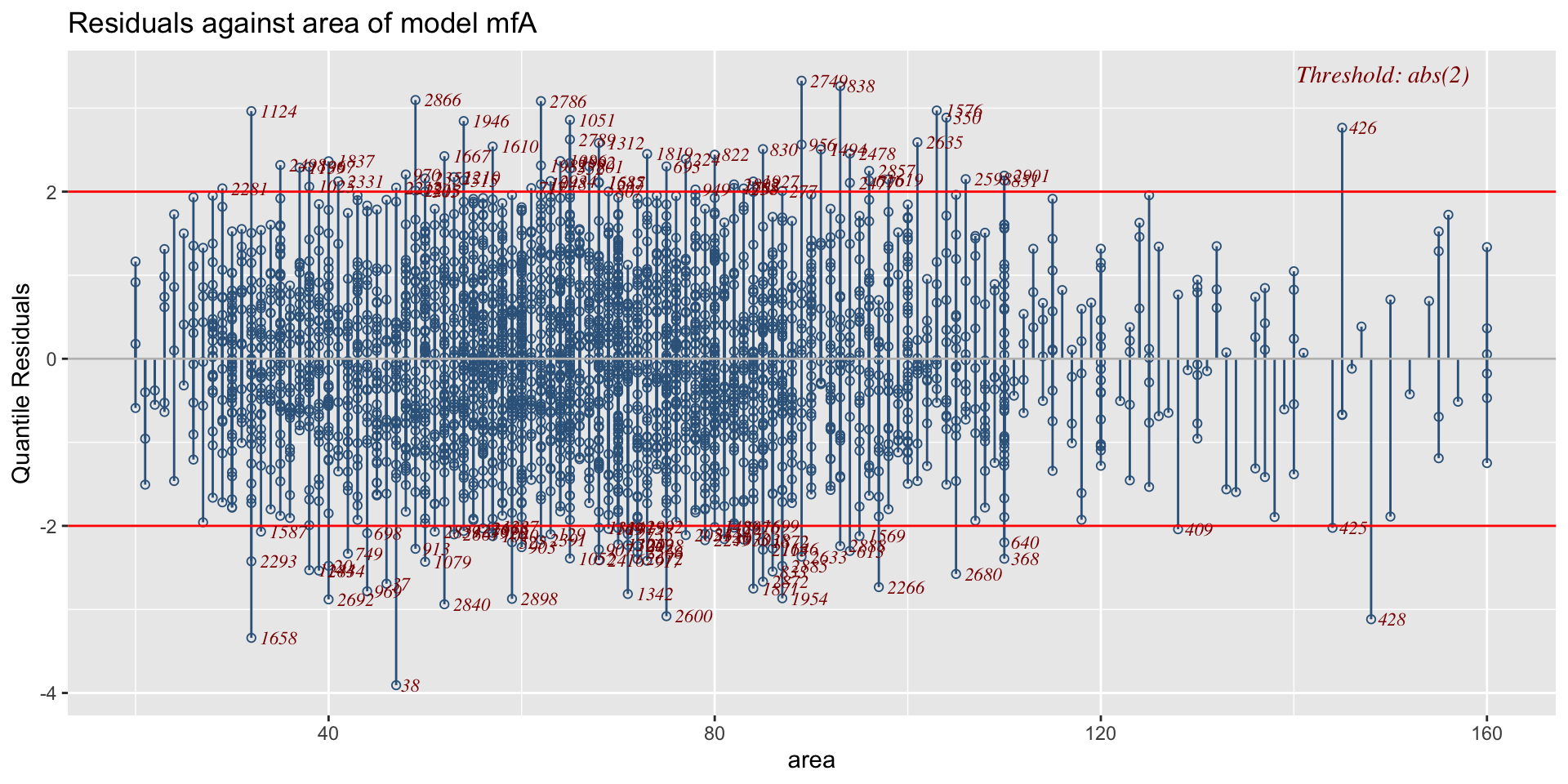
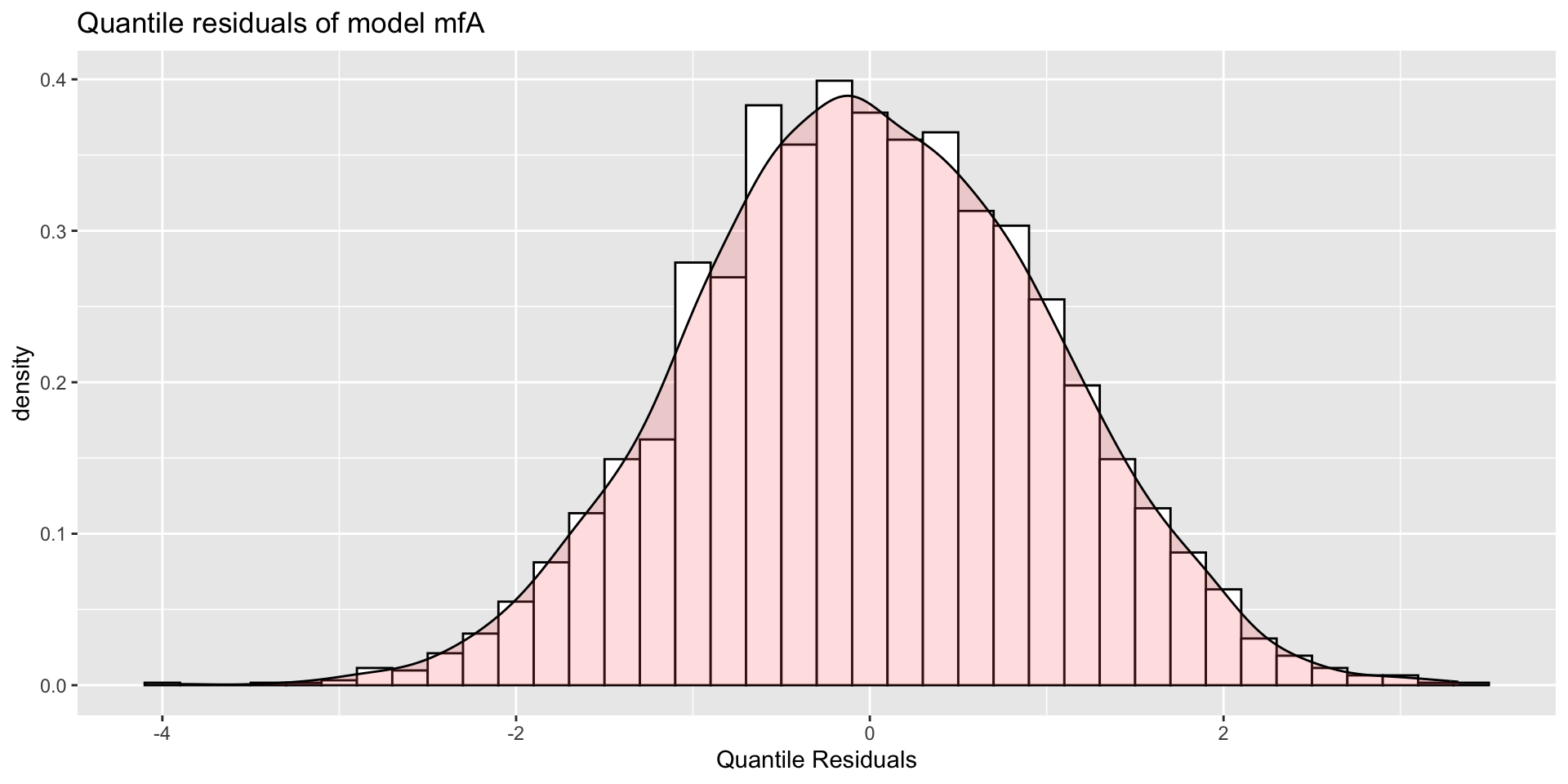
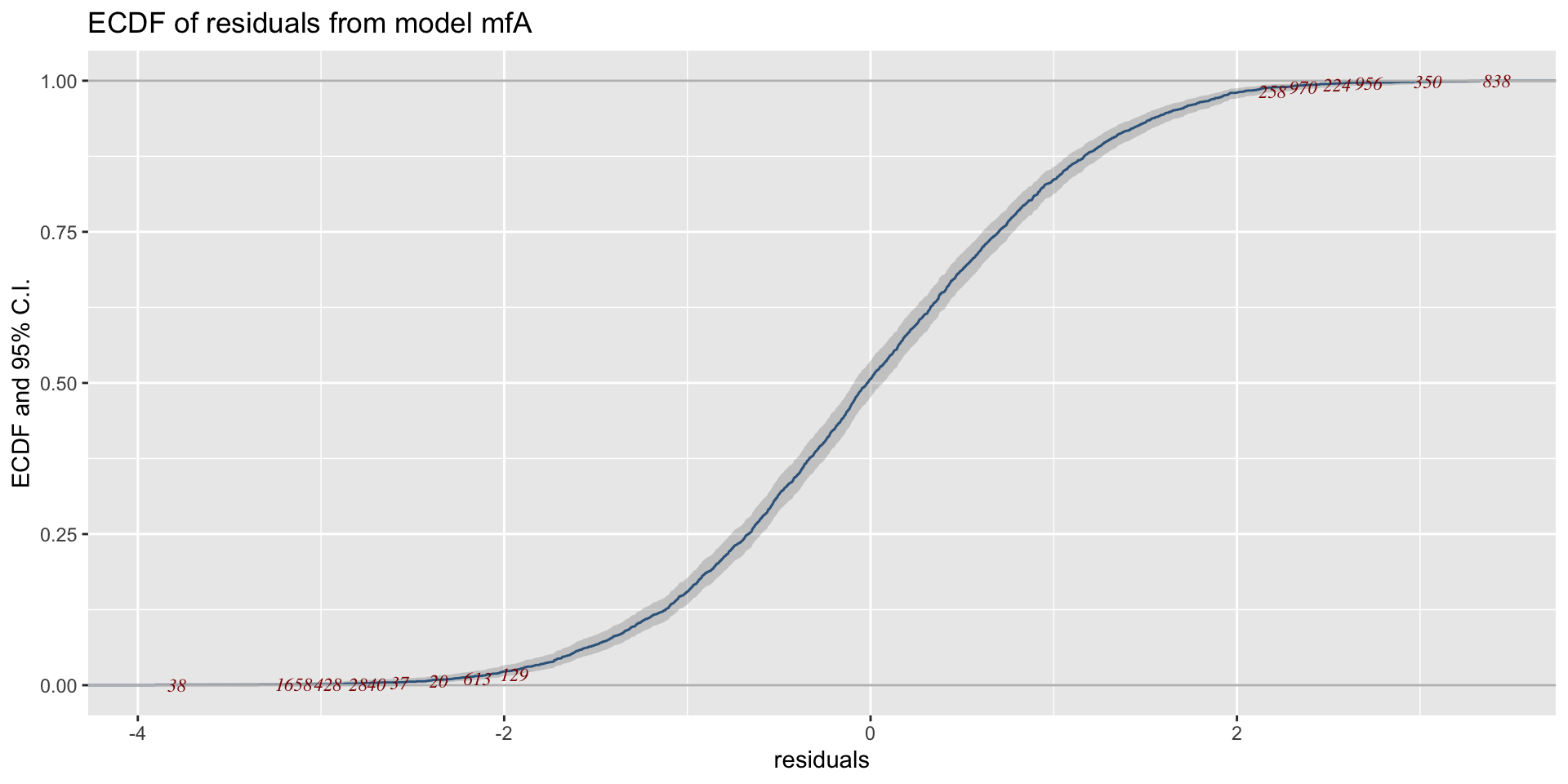
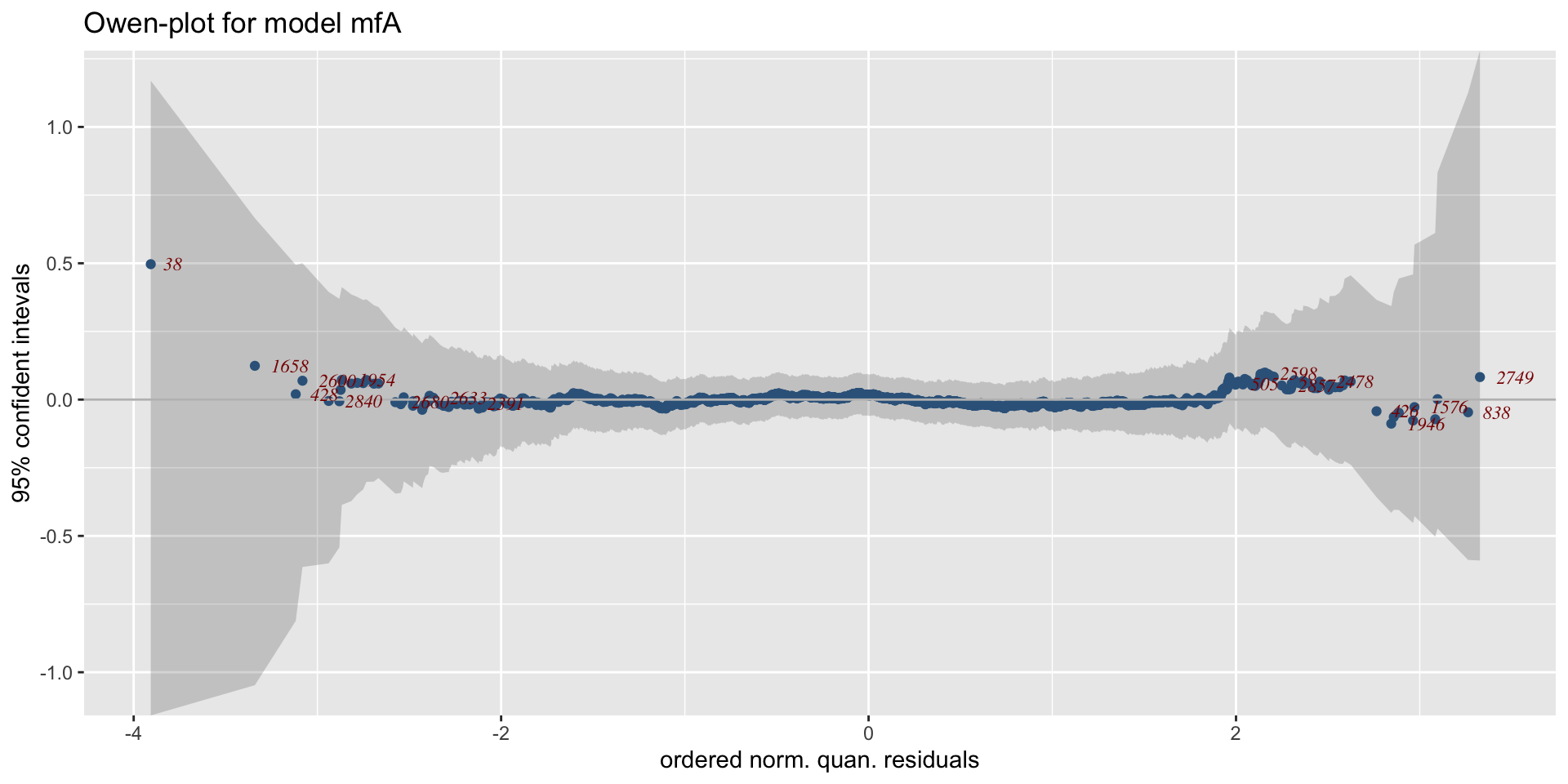
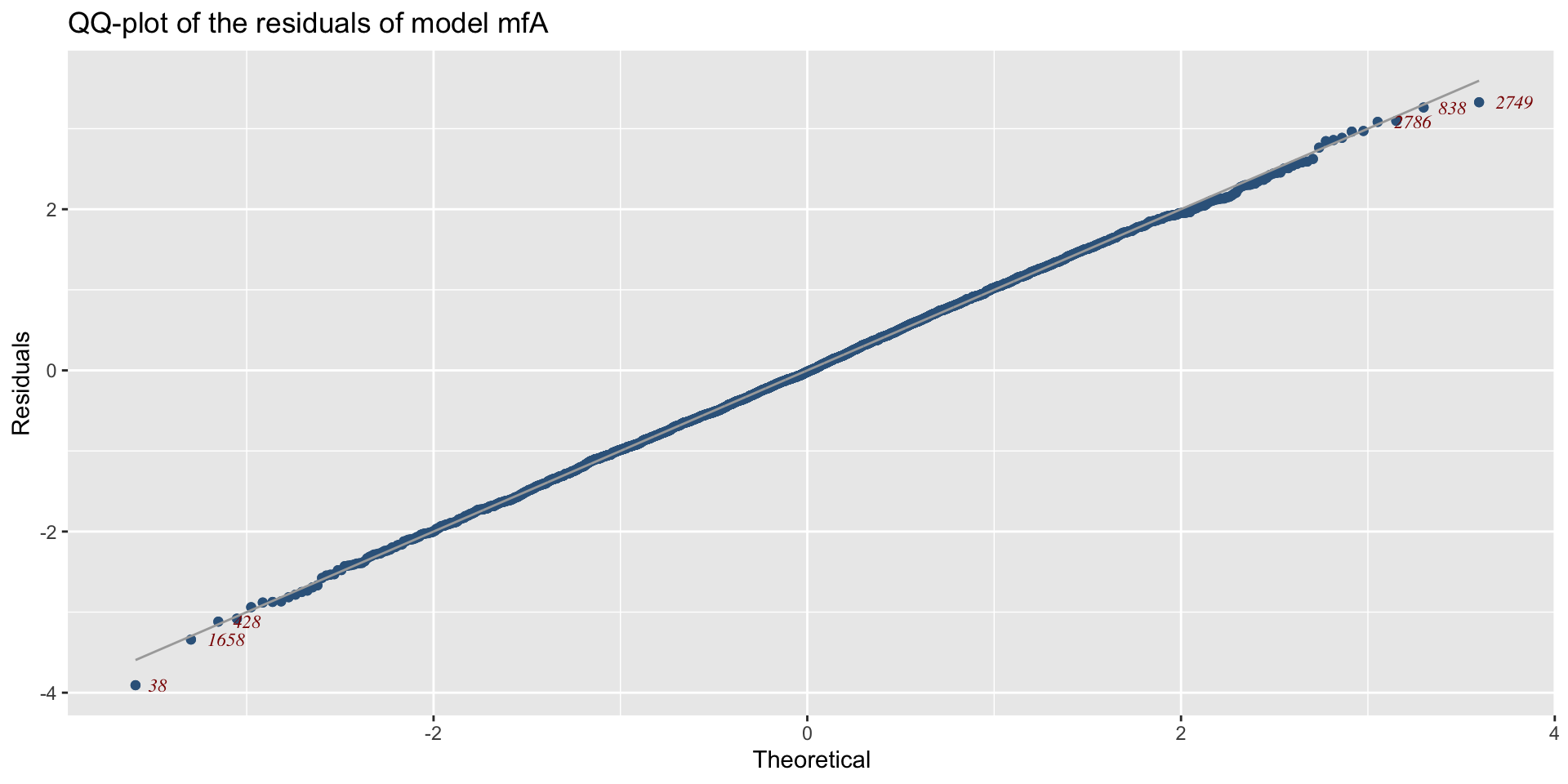
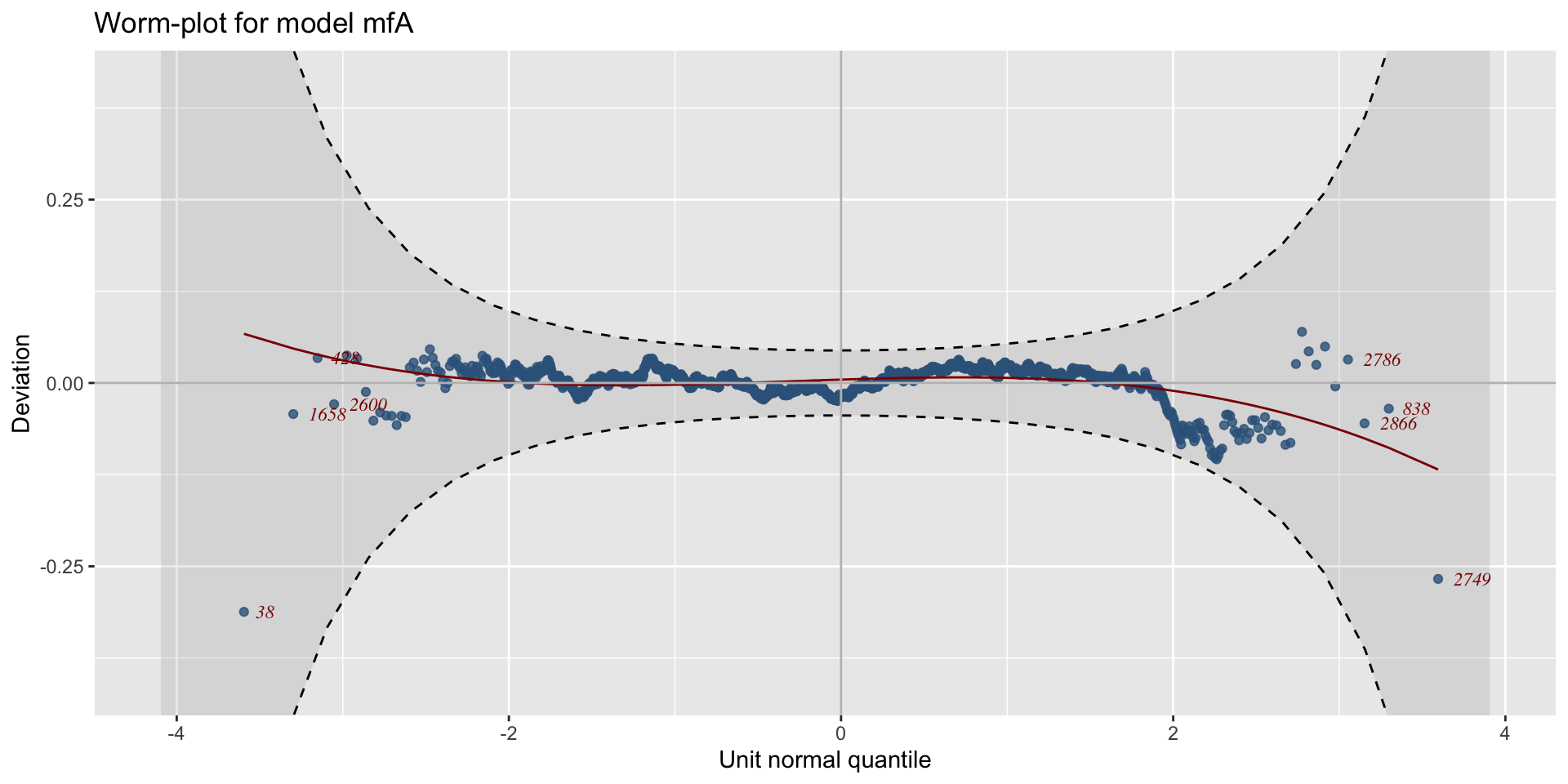
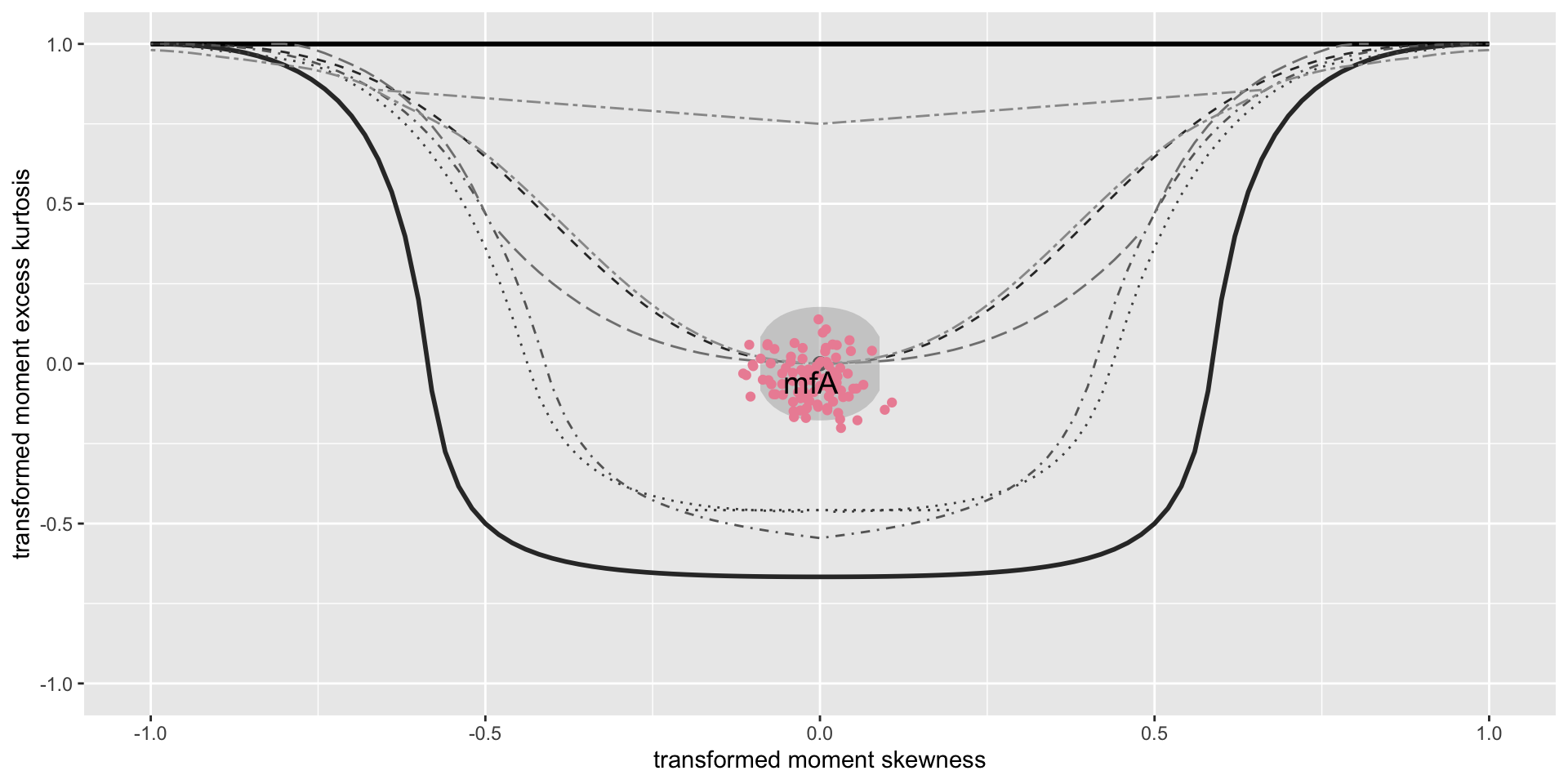
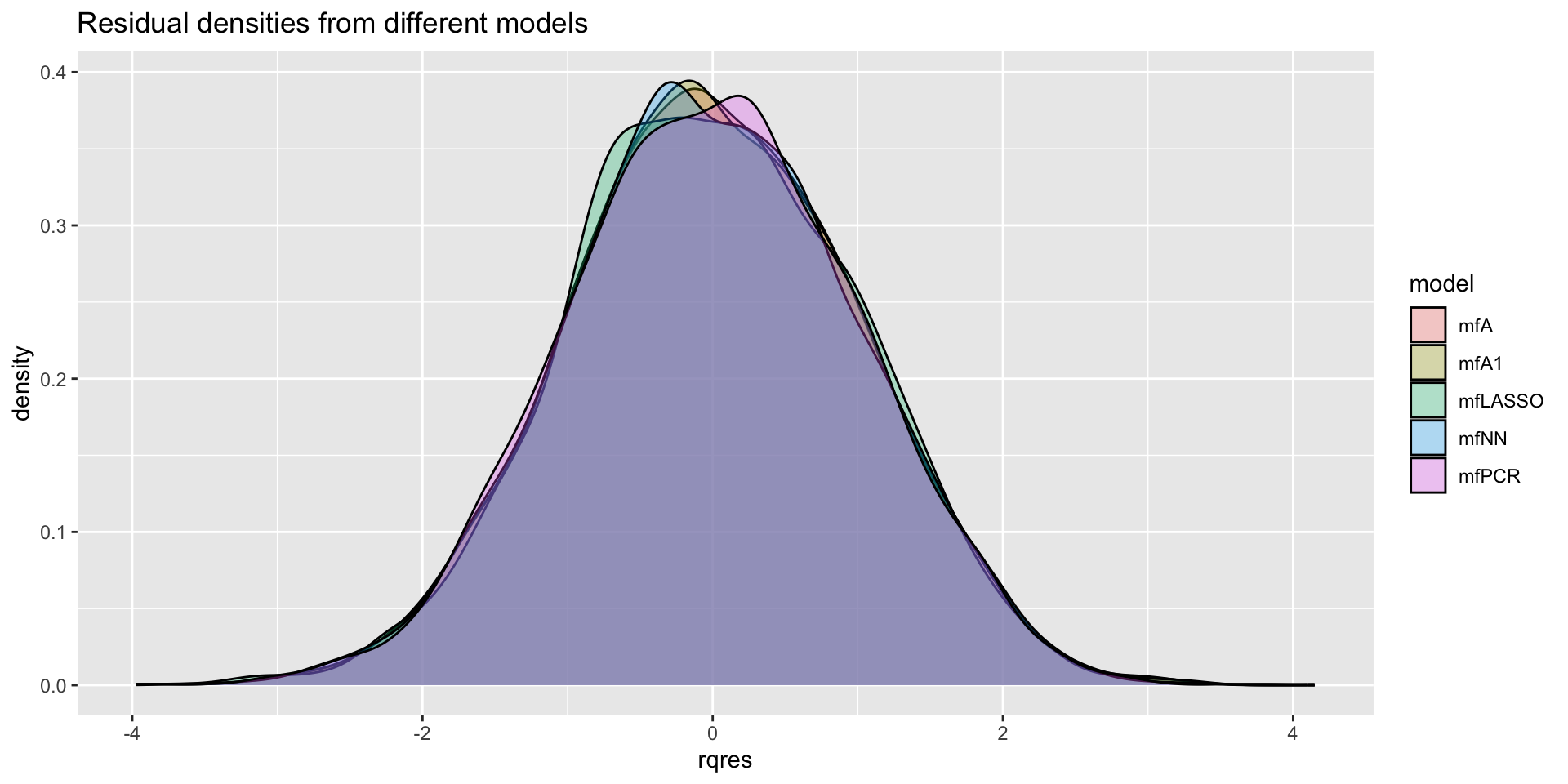
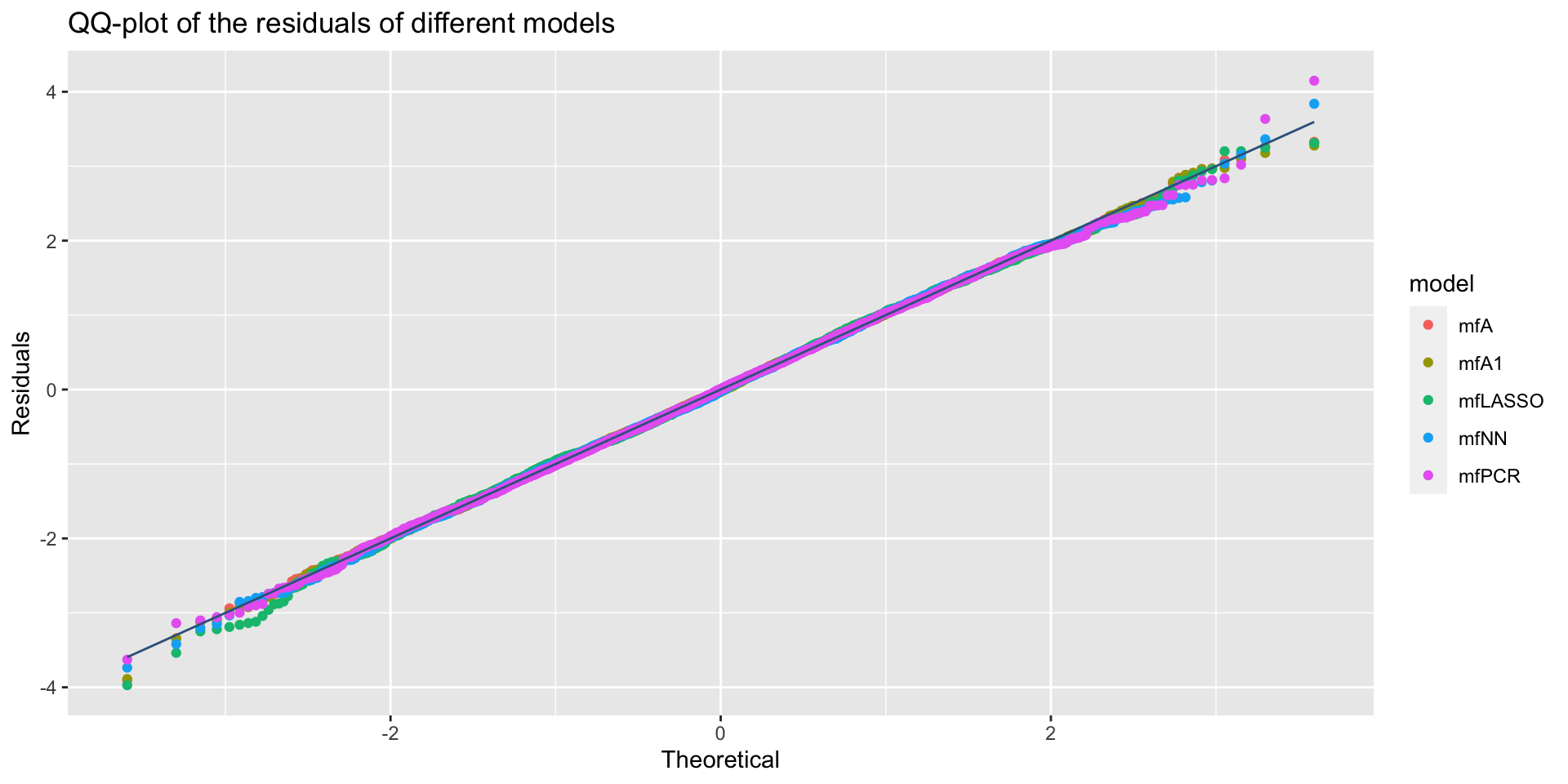
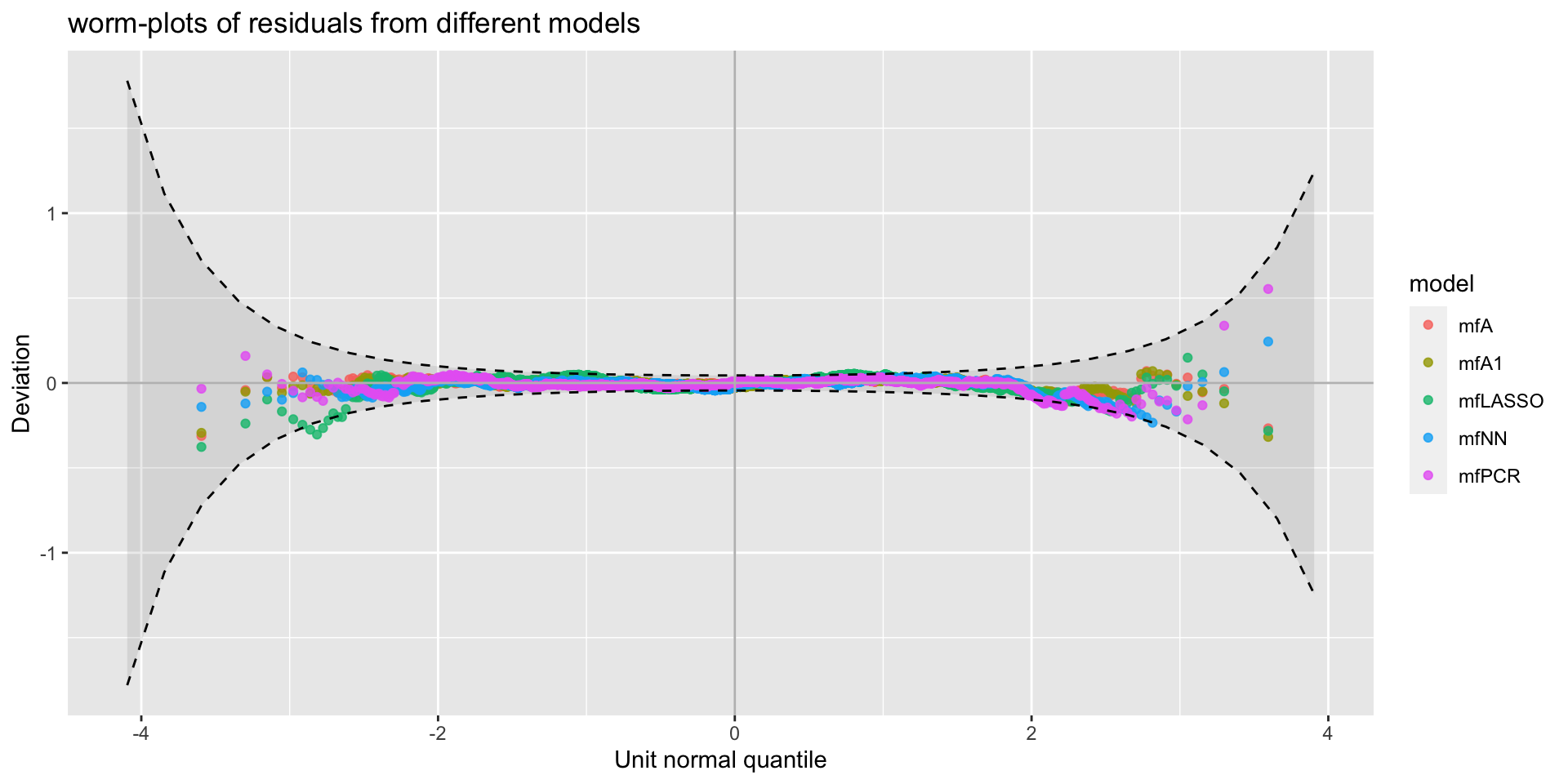
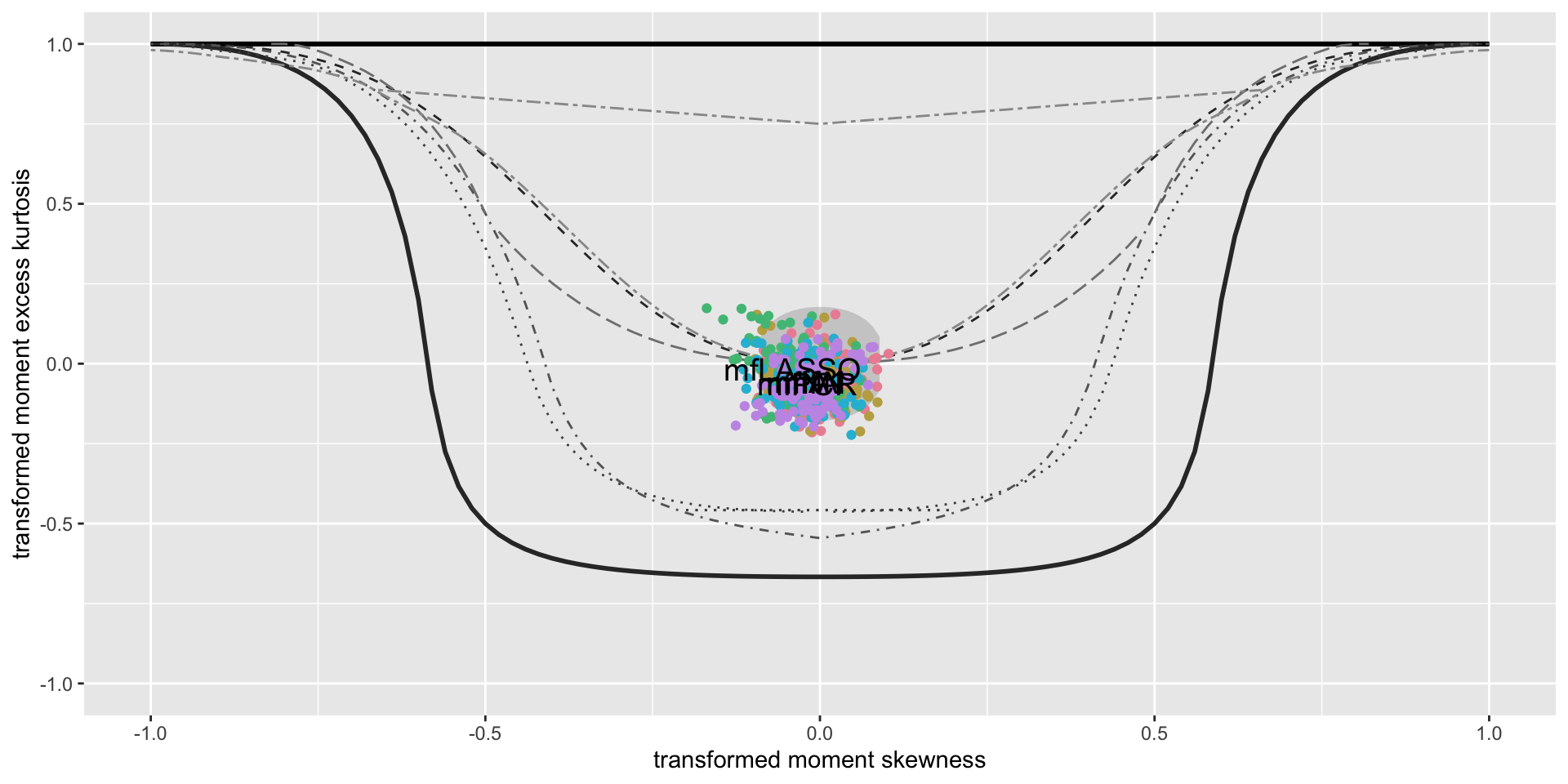
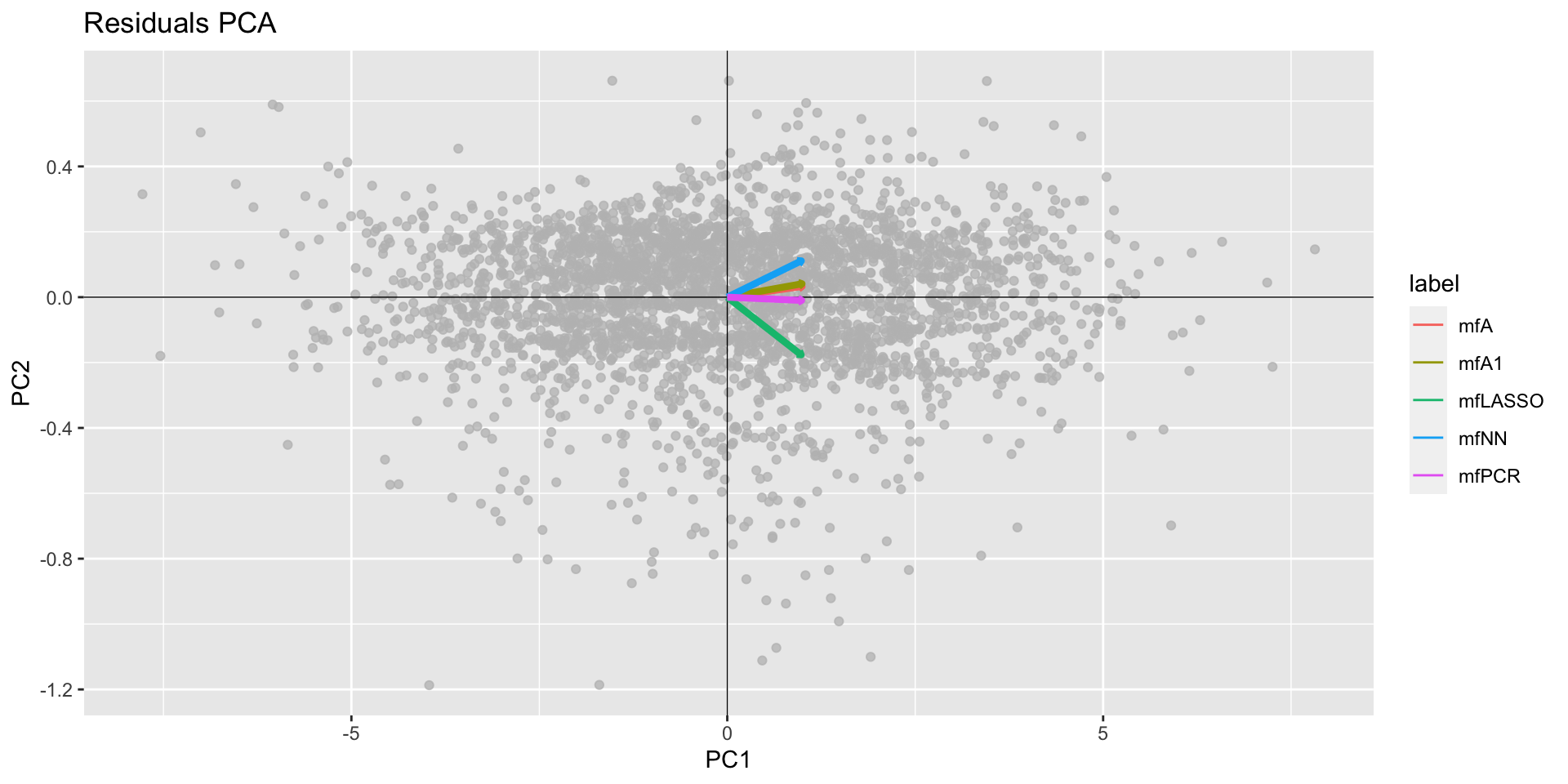
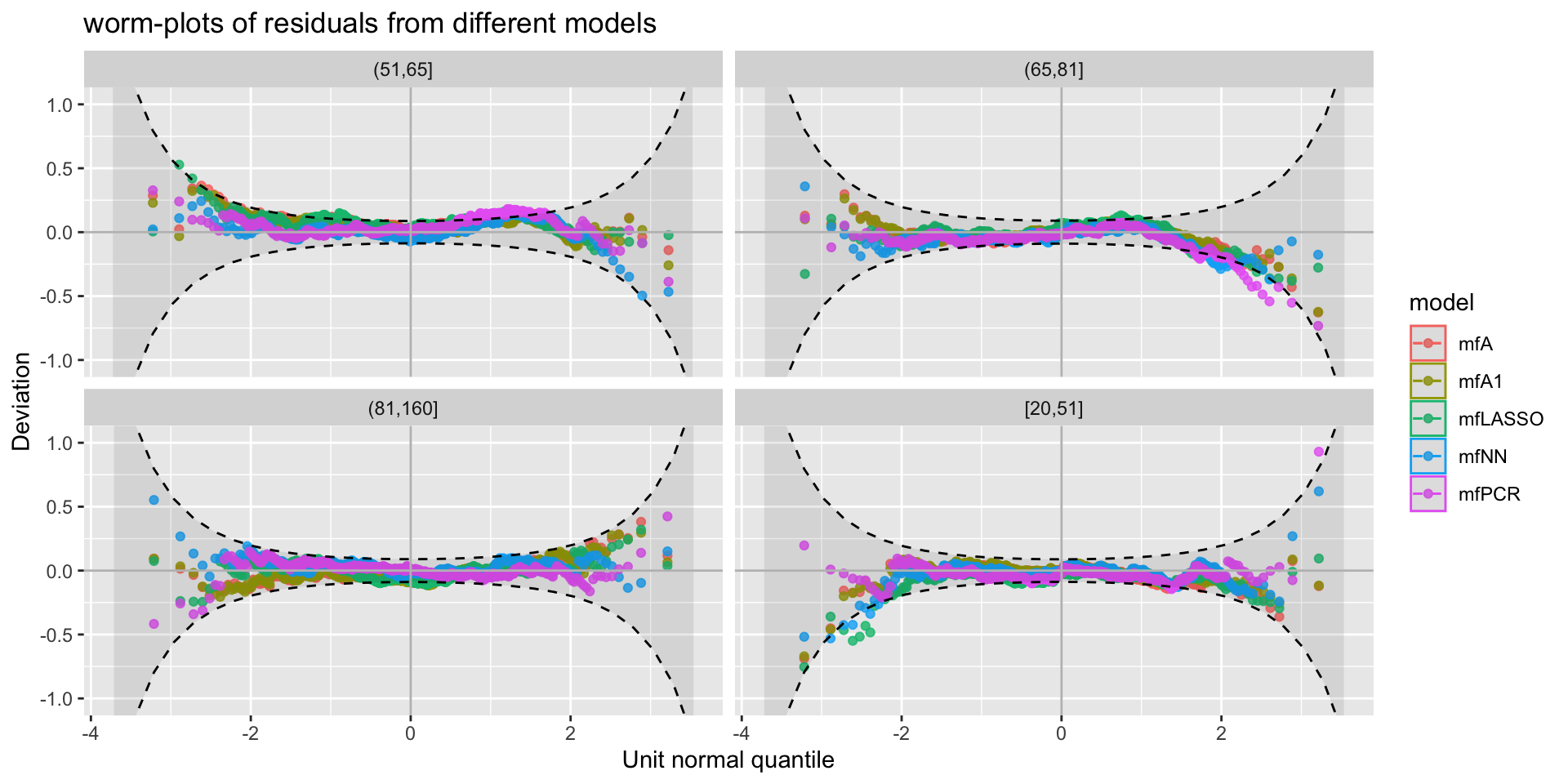
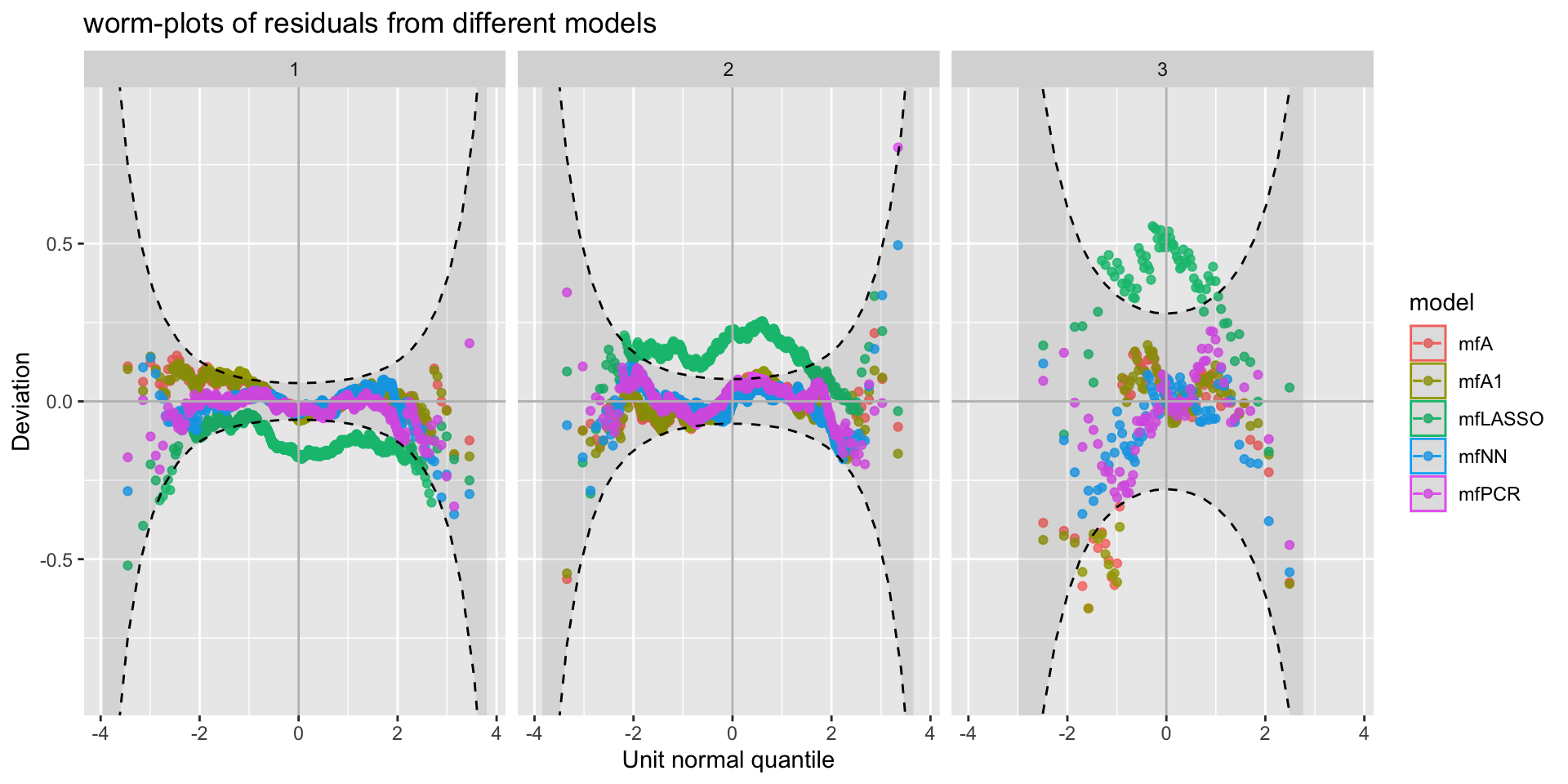
minimum GAIC(k= 2 ) model: mfA1
minimum GAIC(k= 3.84 ) model: mfA1
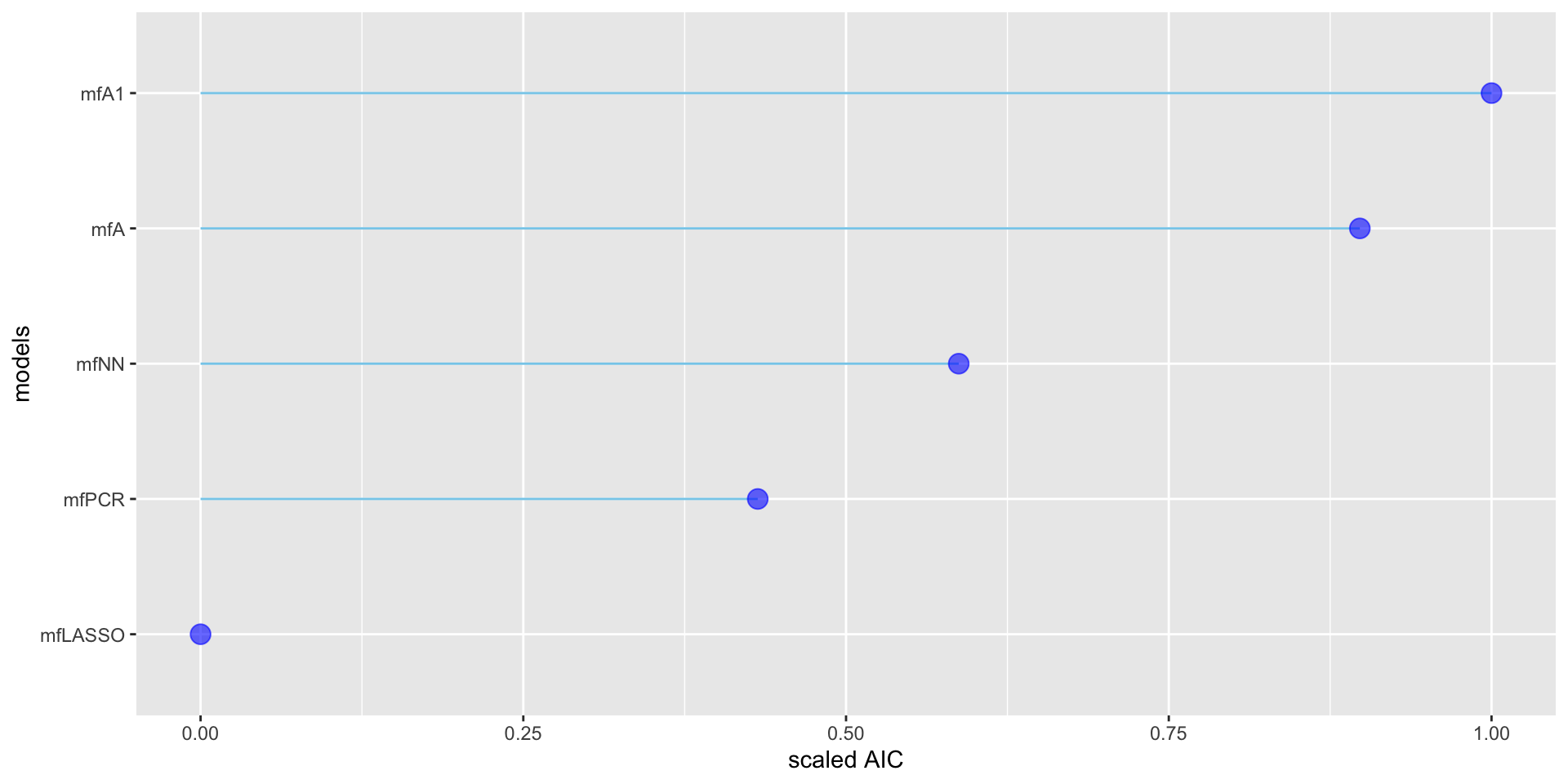
minimum GAIC(k= 8.03 ) model: mfA | df | k=2 | k=3.84 | k=8.03 | |
|---|---|---|---|---|
| mfA | 23 | 38169.3 | 38211.6 | 38308.0 |
| mfA1 | 27 | 38156.1 | 38206.0 | 38319.8 |
| mfLASSO | 23 | 38285.4 | 38327.7 | 38424.1 |
| mfNN | 134 | 38209.4 | 38456.0 | 39017.5 |
| mfPCR | 68 | 38229.6 | 38354.7 | 38639.6 |


 The Books
The Books