library(gamlss2)
library(gamlss.ggplots)
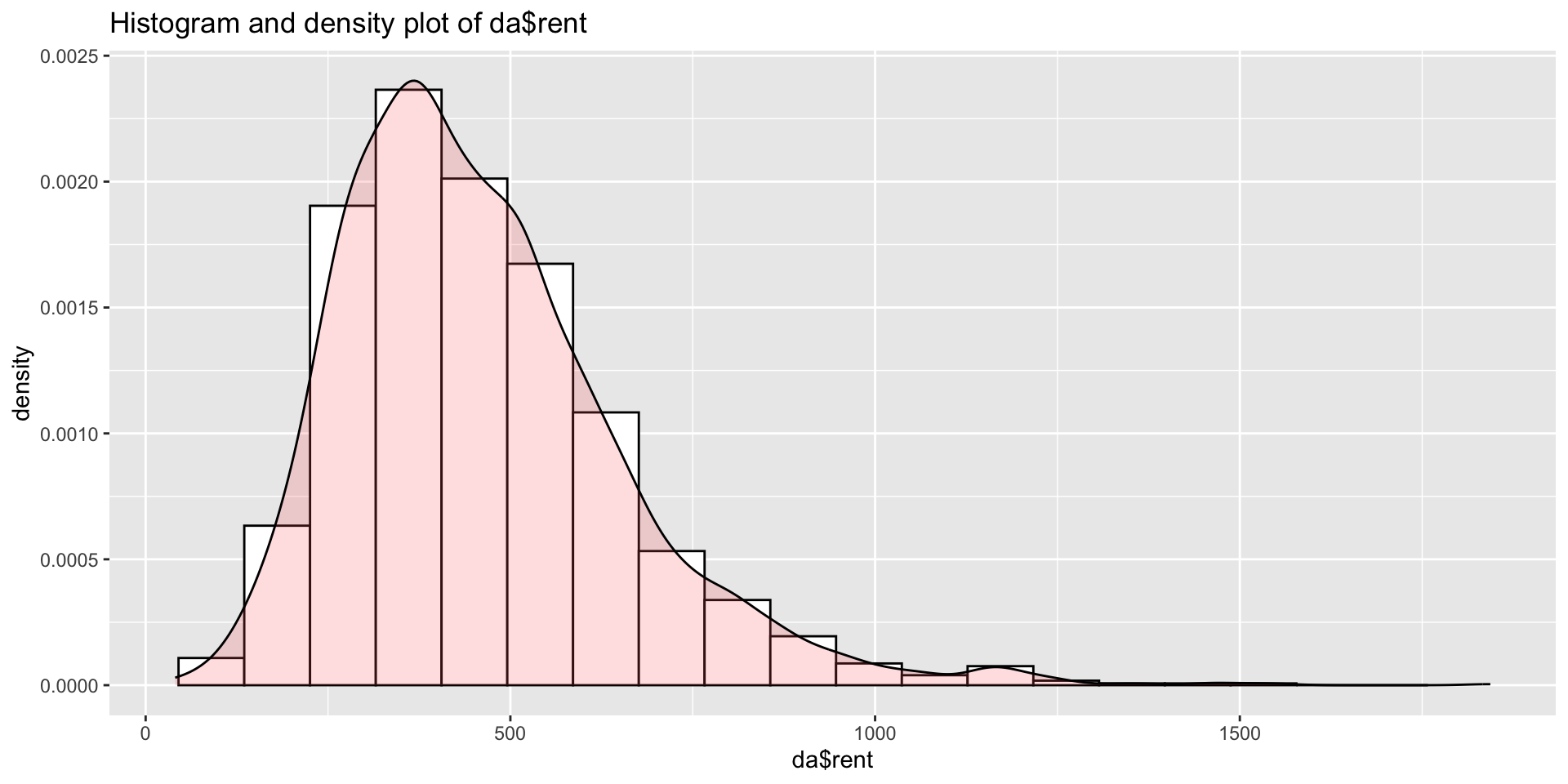
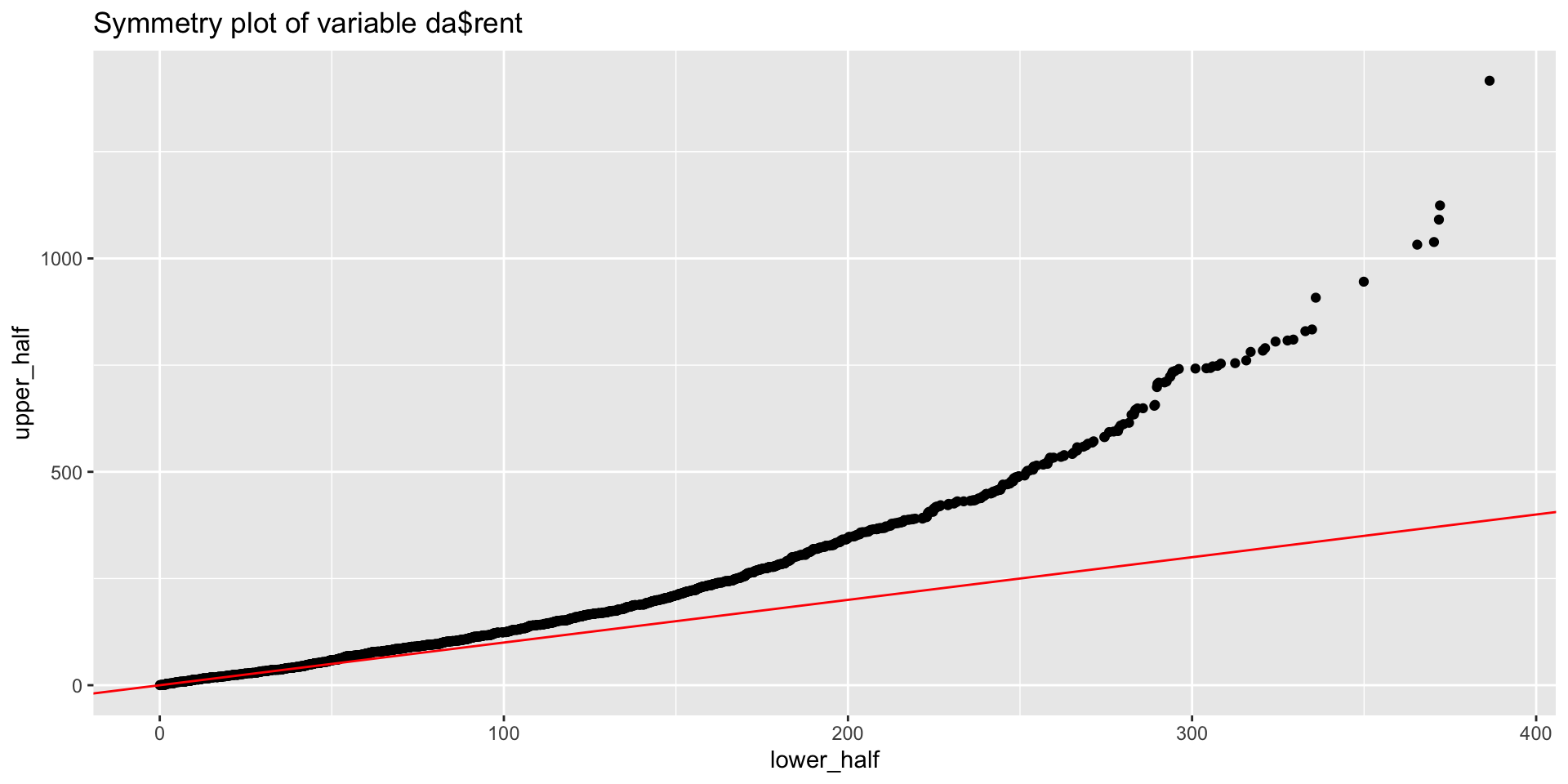
da <- rent99[,-c(2,9)]
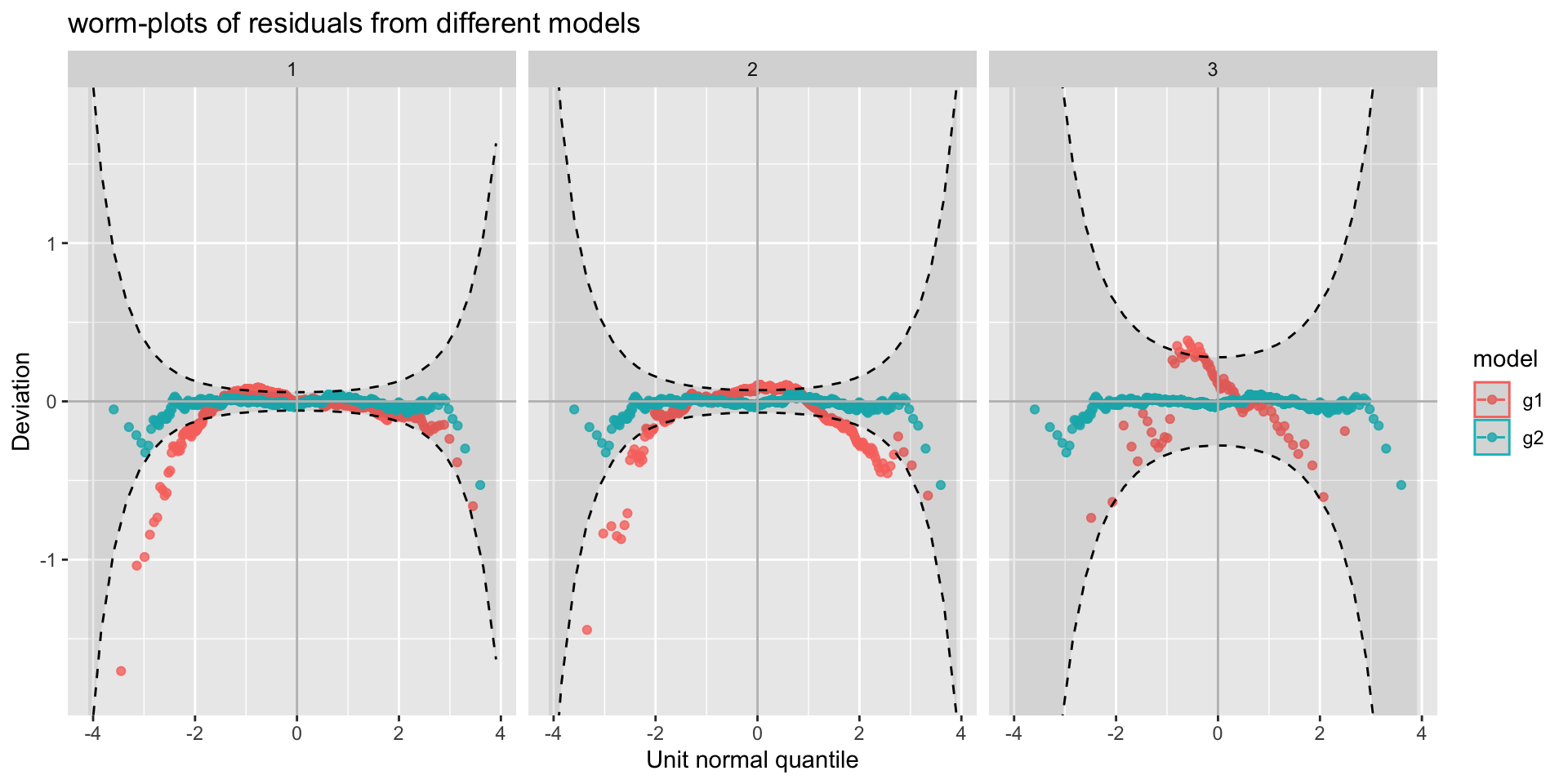
g1 <- gamlss2(rent~s(area)+s(yearc)+bath+location+kitchen+cheating|
s(area)+s(yearc)+bath+location+kitchen+cheating, data=da, family=GA )
GAMLSS-RS iteration 1: Global Deviance = 38249.9521 eps = 0.129707
GAMLSS-RS iteration 2: Global Deviance = 38205.9938 eps = 0.001149
GAMLSS-RS iteration 3: Global Deviance = 38205.9065 eps = 0.000002
g2 <- gamlss2(rent~s(area)+s(yearc)+bath+location+kitchen+cheating|
s(area)+s(yearc)+bath+location+kitchen+cheating, data=da, family=BCTo )
GAMLSS-RS iteration 1: Global Deviance = 38170.7368 eps = 0.290582
GAMLSS-RS iteration 2: Global Deviance = 38135.3004 eps = 0.000928
GAMLSS-RS iteration 3: Global Deviance = 38133.3037 eps = 0.000052
GAMLSS-RS iteration 4: Global Deviance = 38132.6789 eps = 0.000016
GAMLSS-RS iteration 5: Global Deviance = 38132.4637 eps = 0.000005
AIC df
g2 38196.95 32.24370
g1 38267.22 30.65513
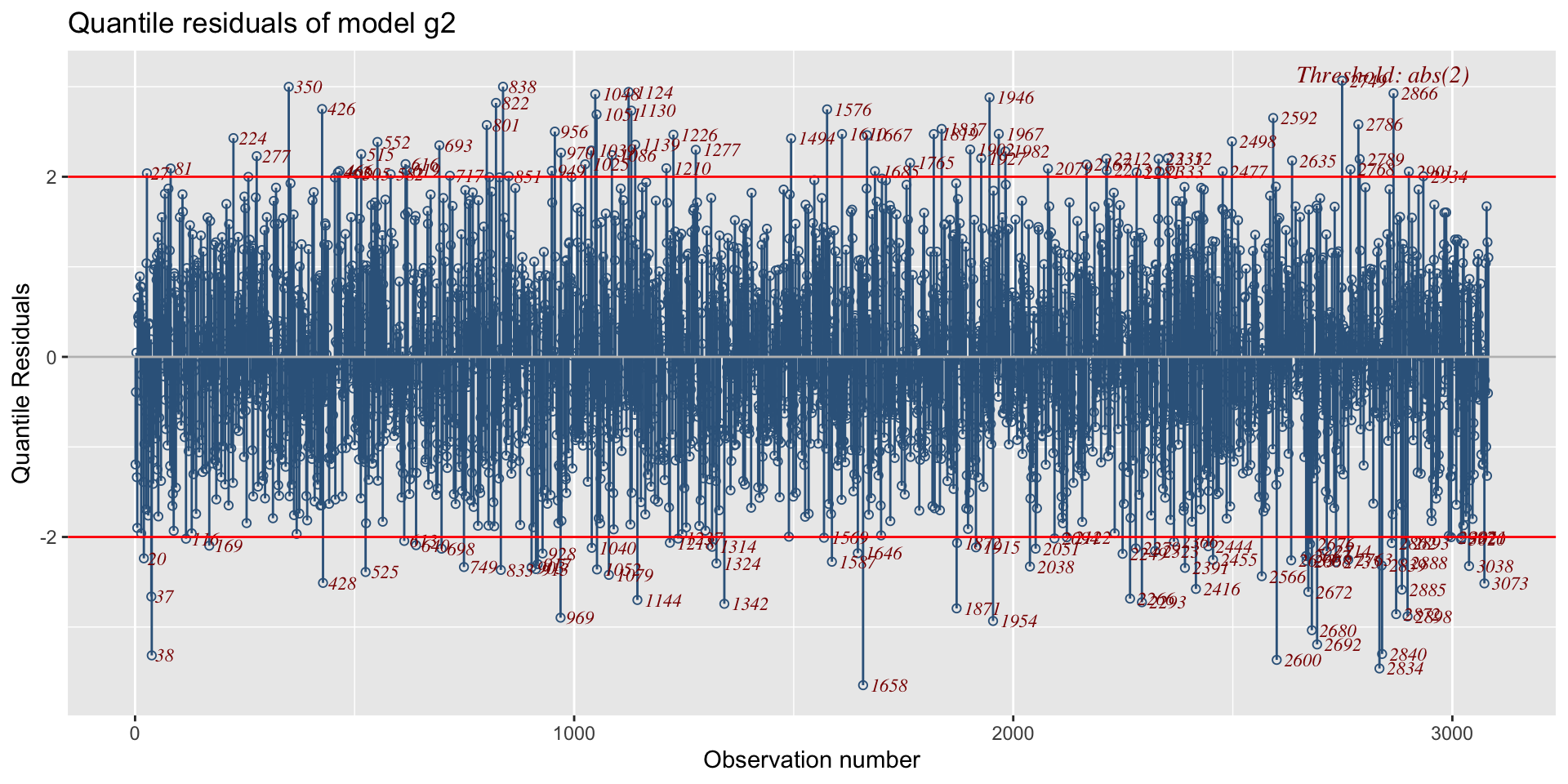
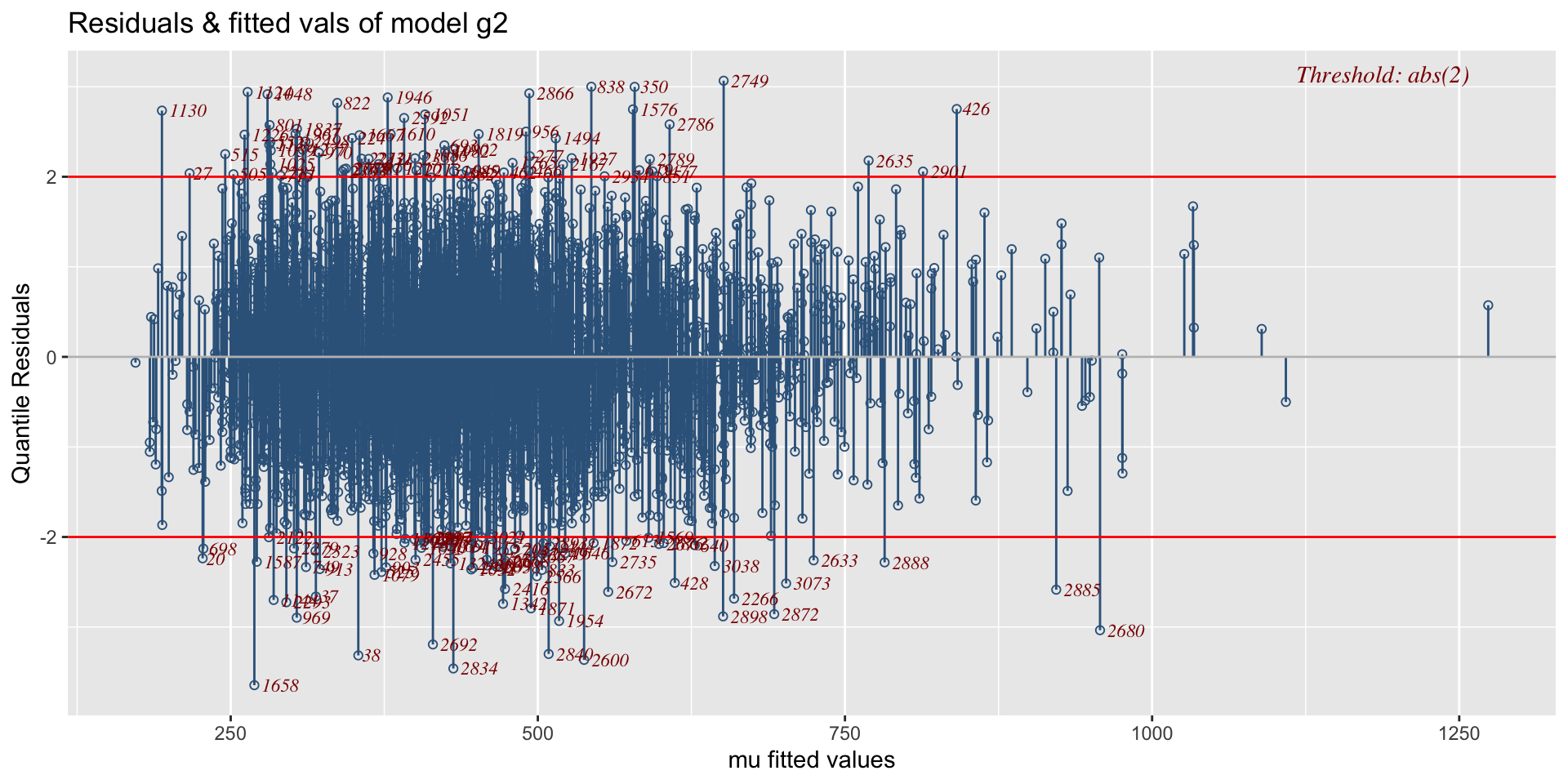
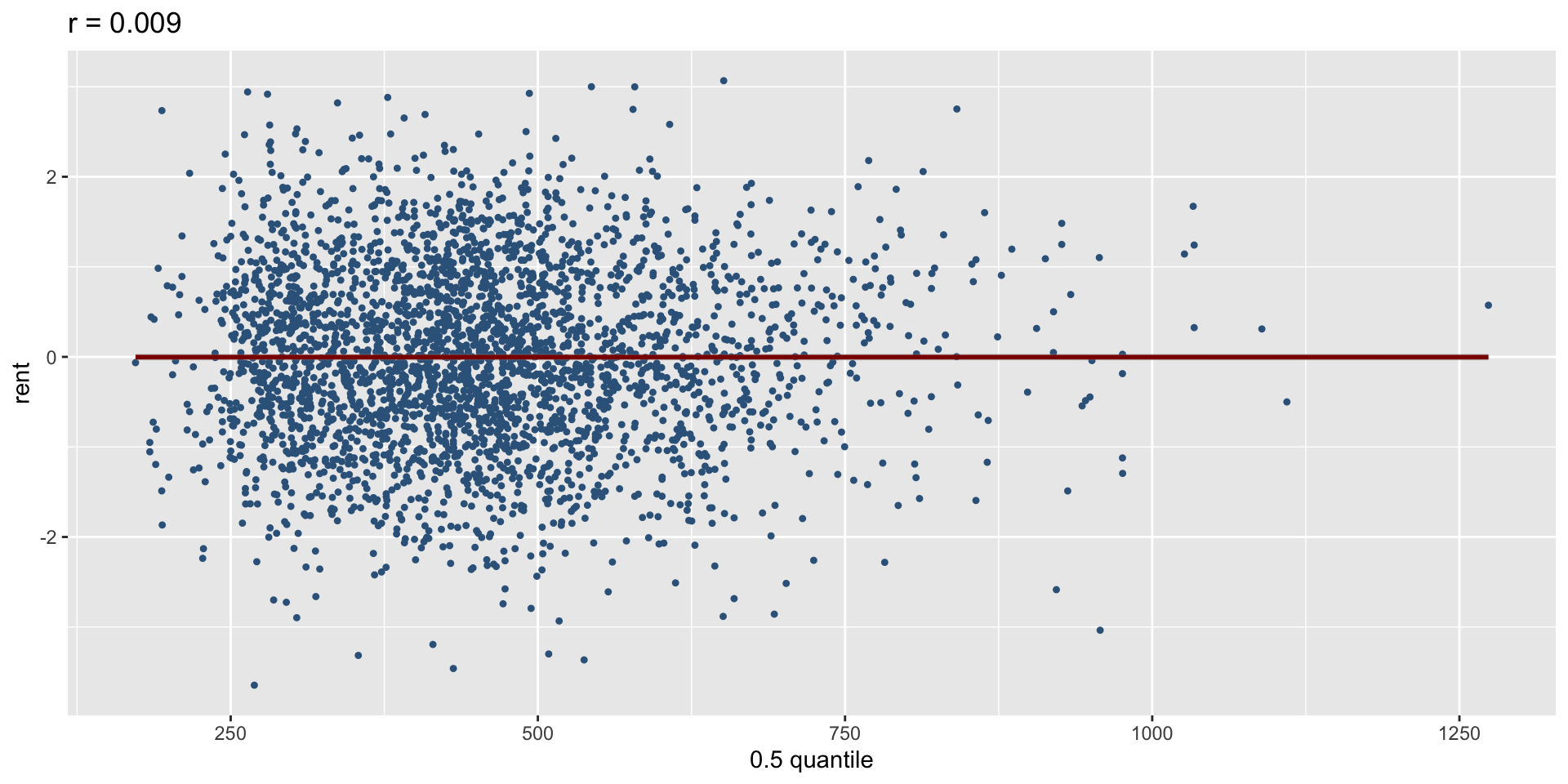
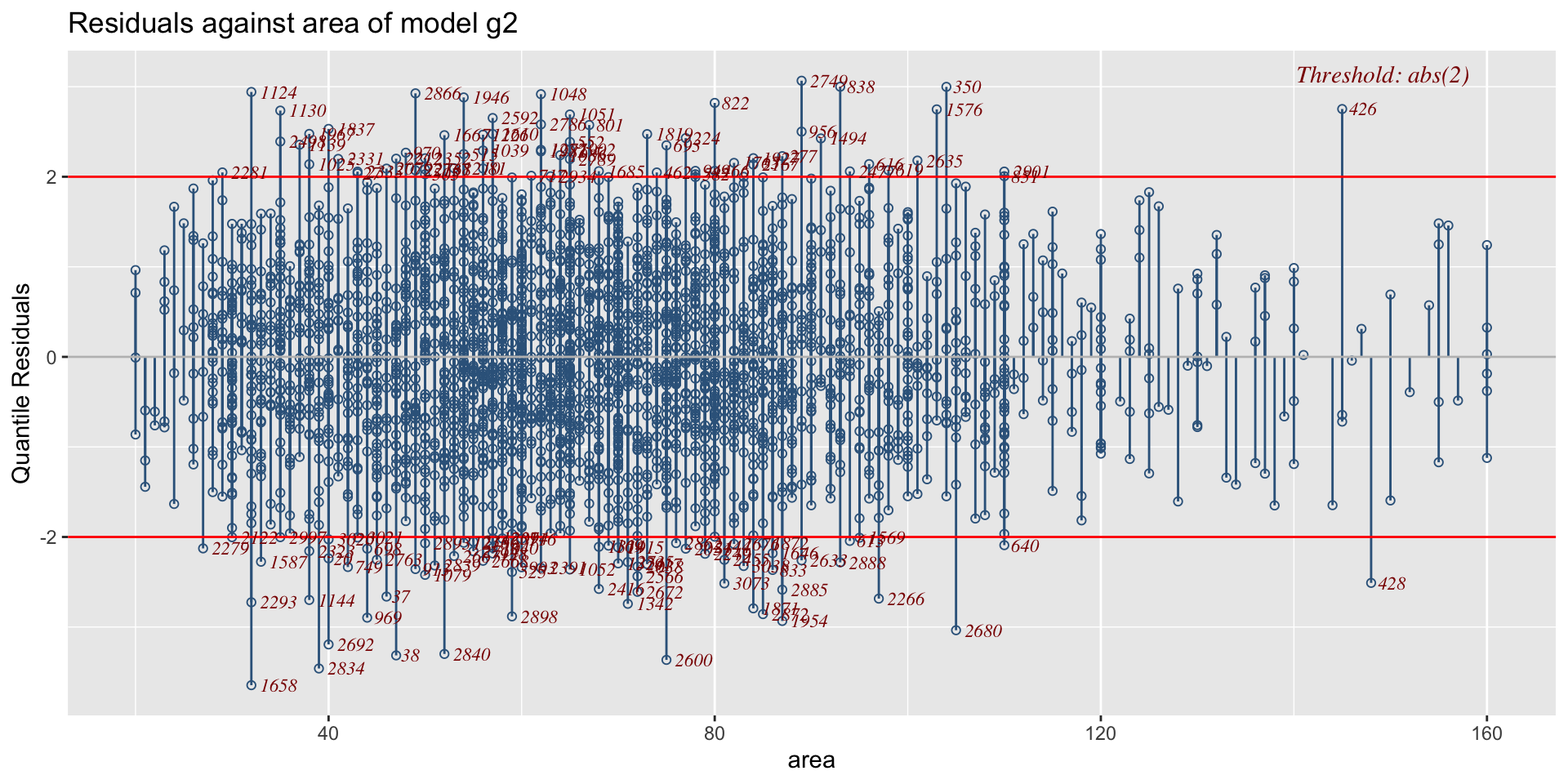
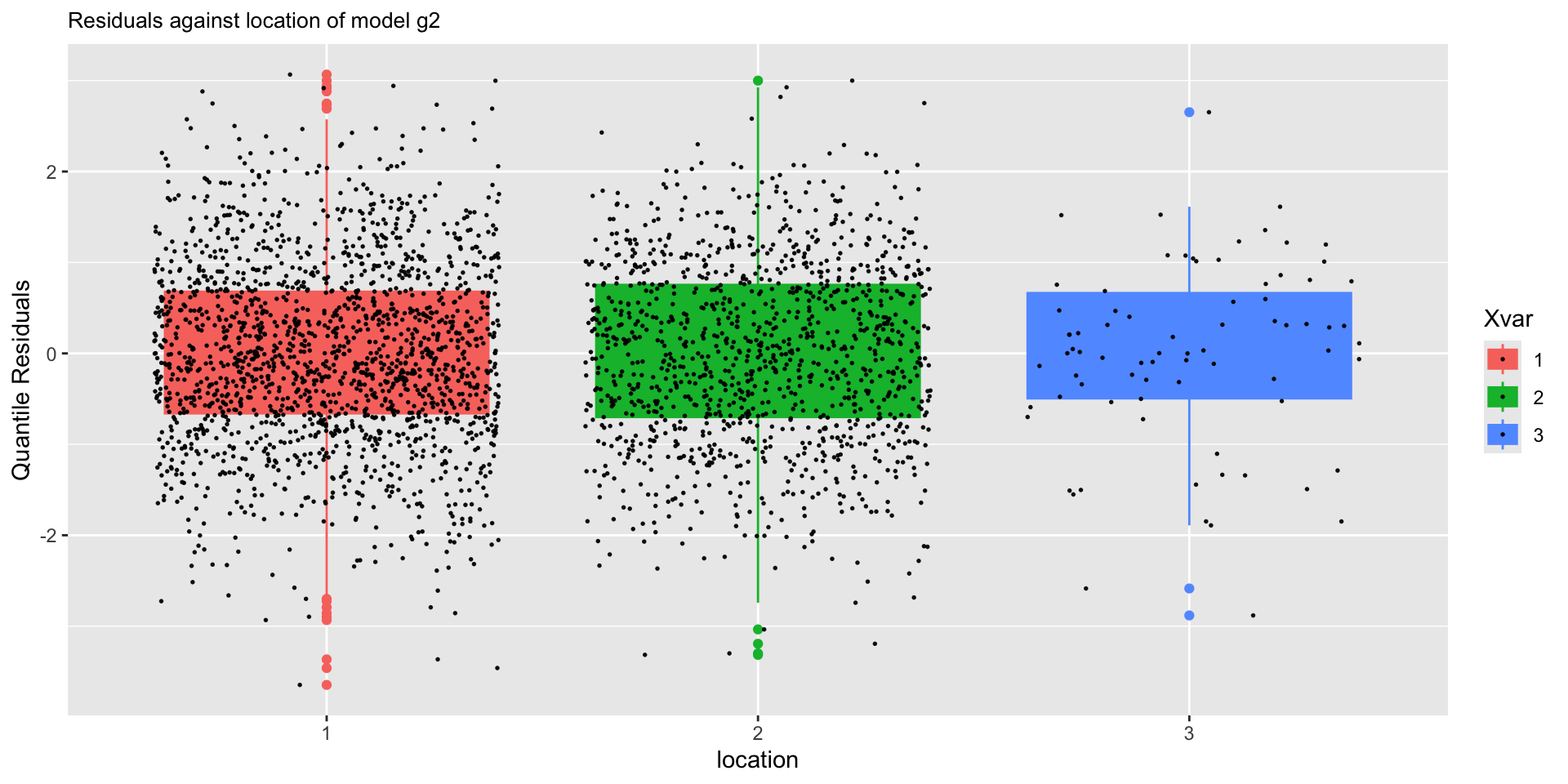
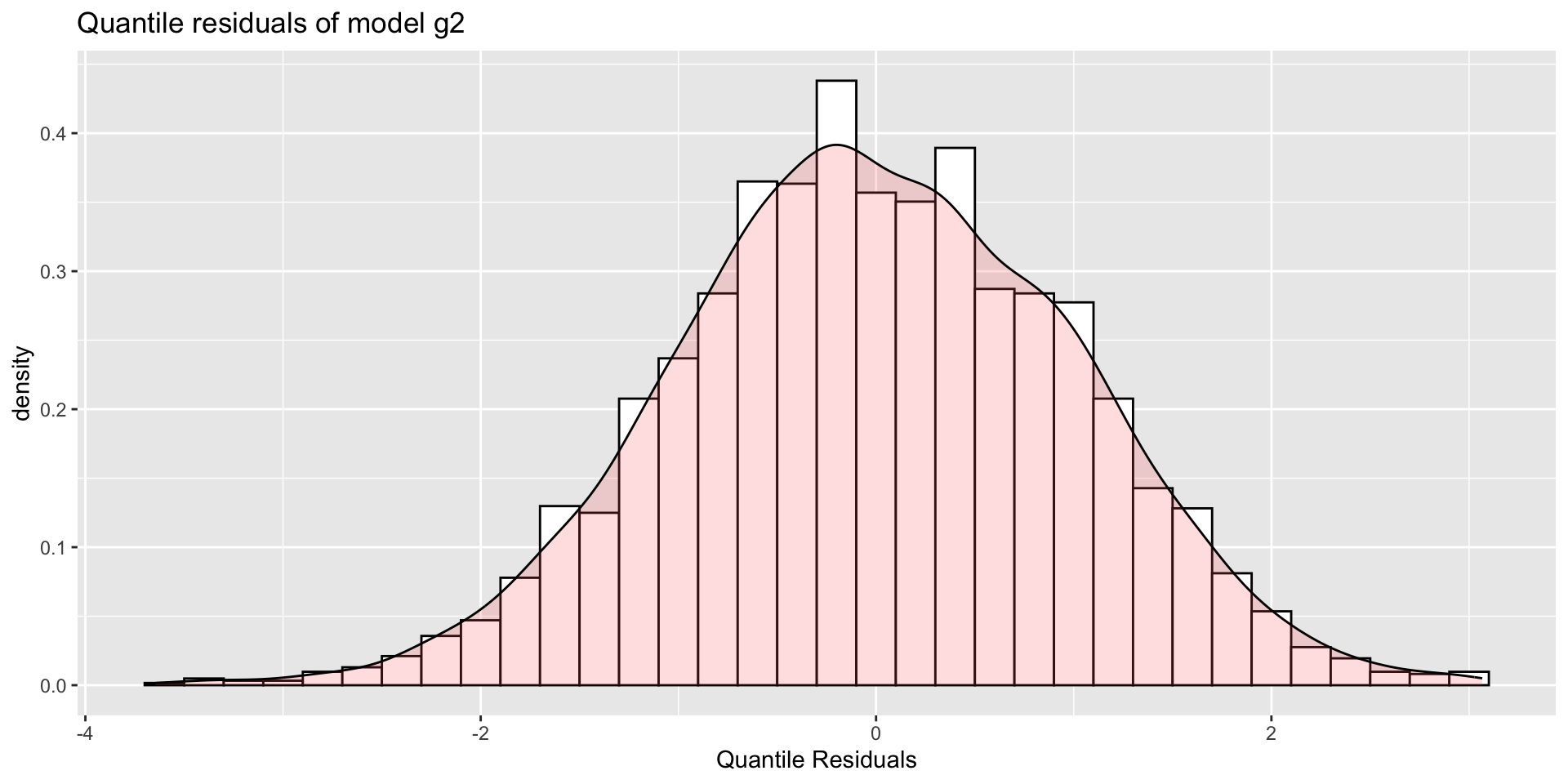
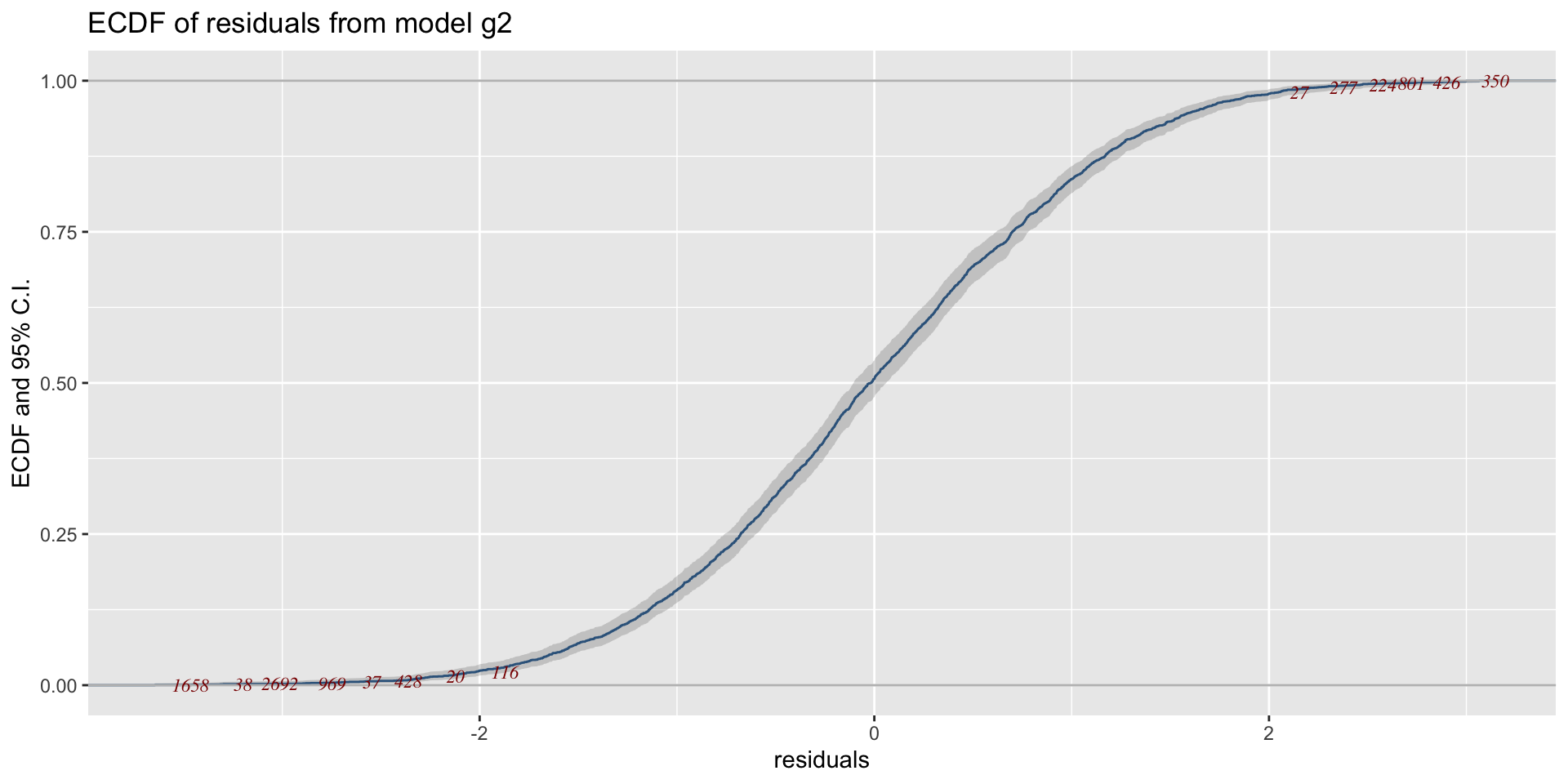
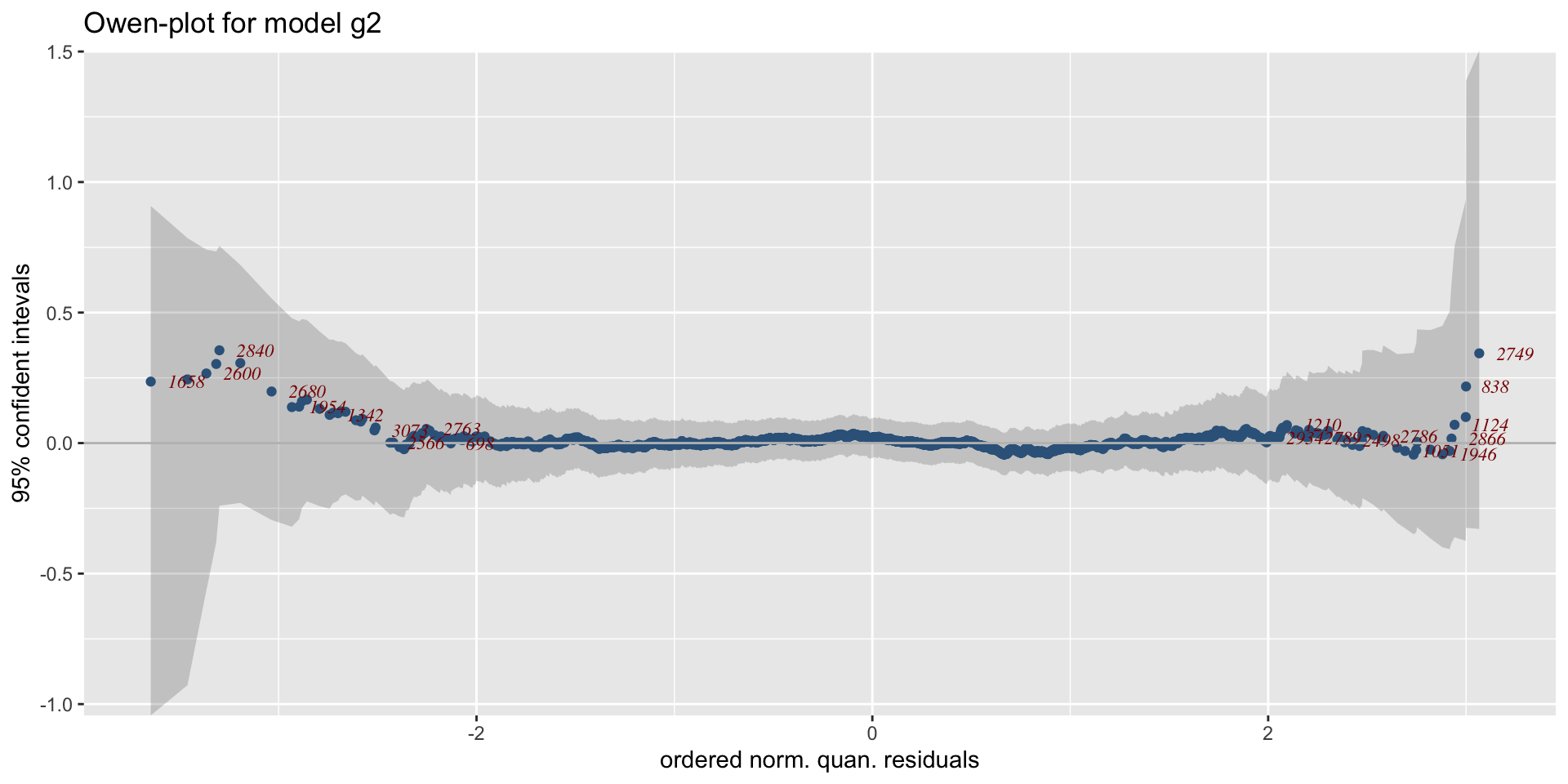
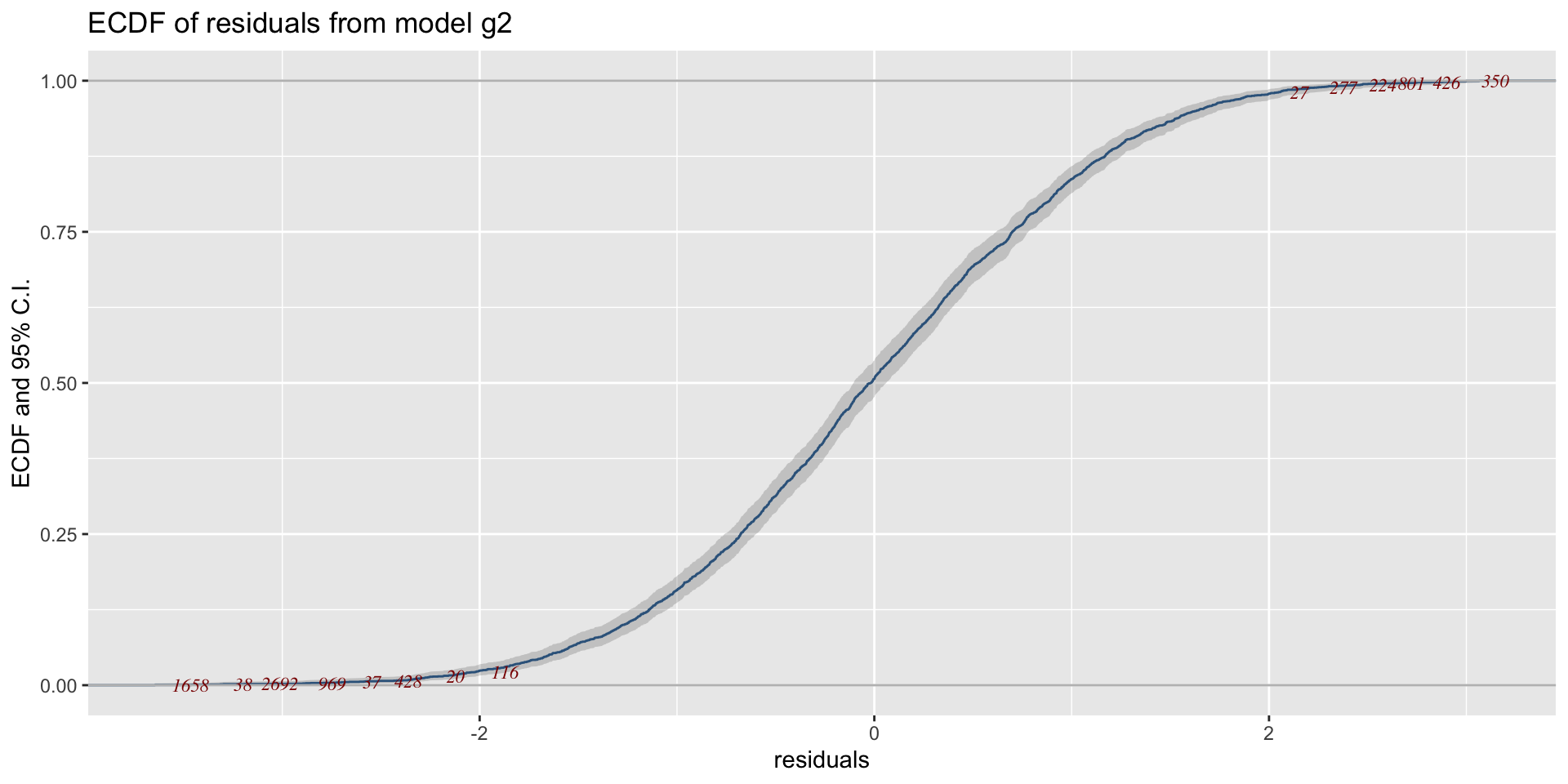
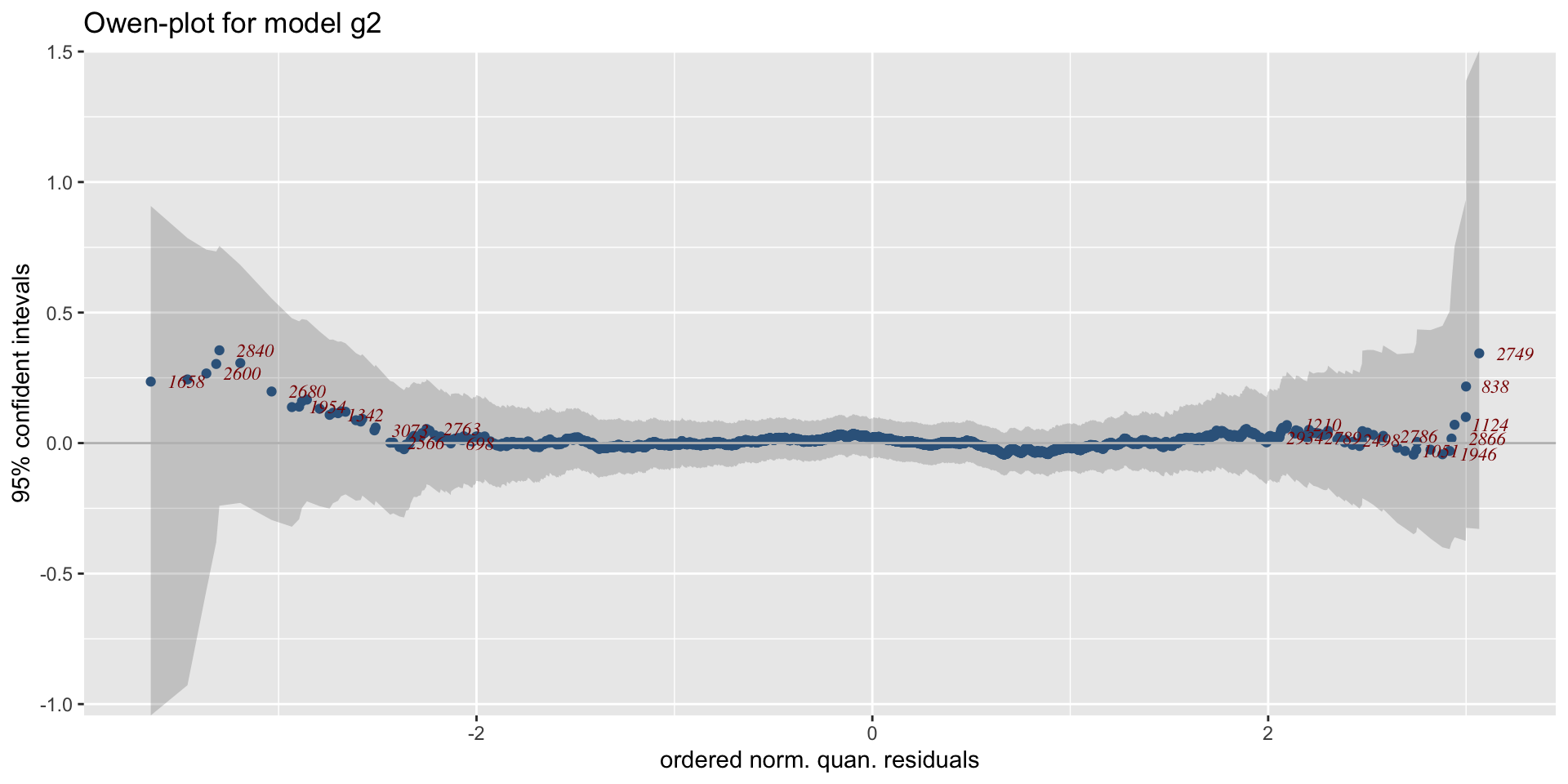
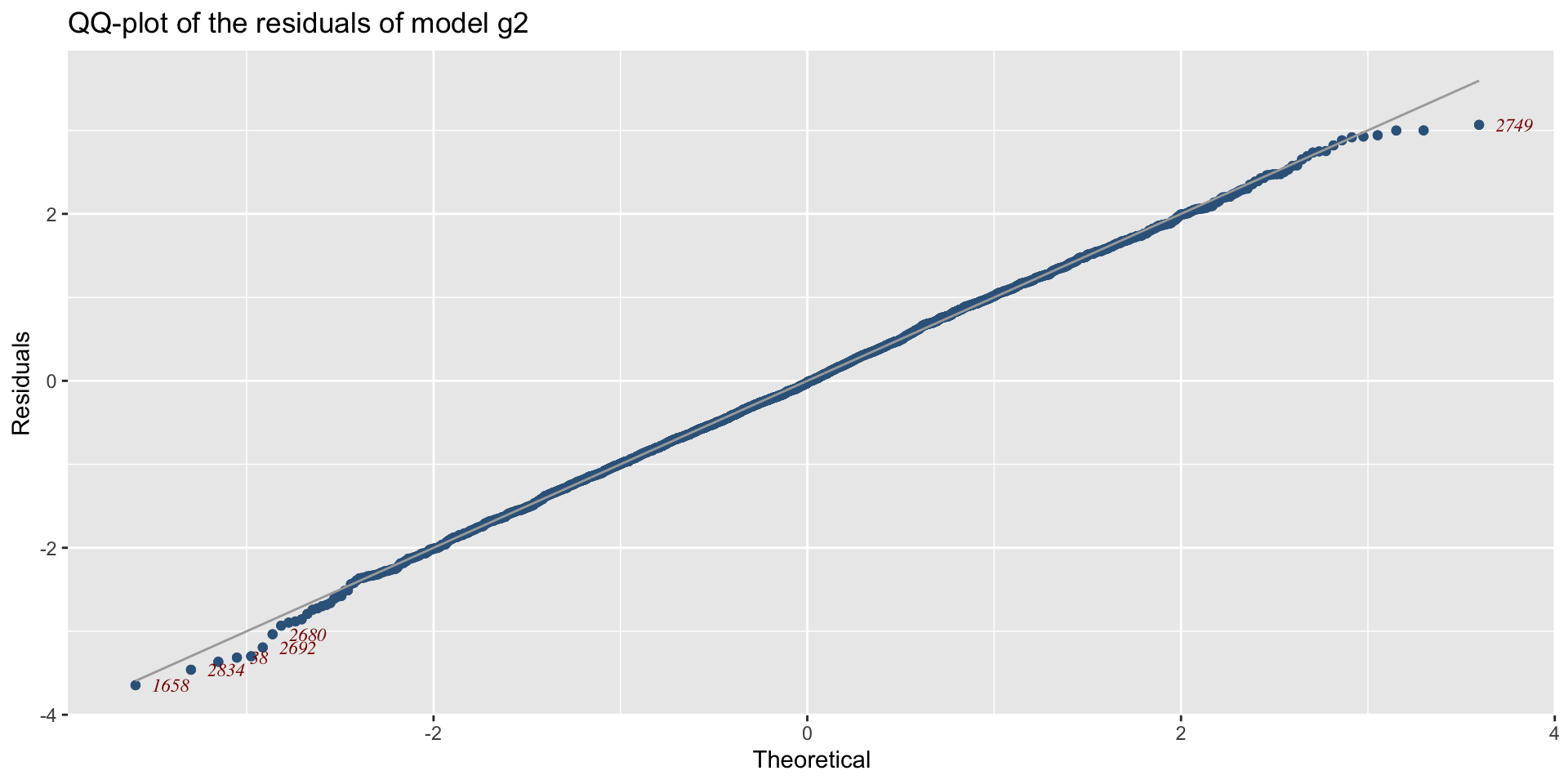
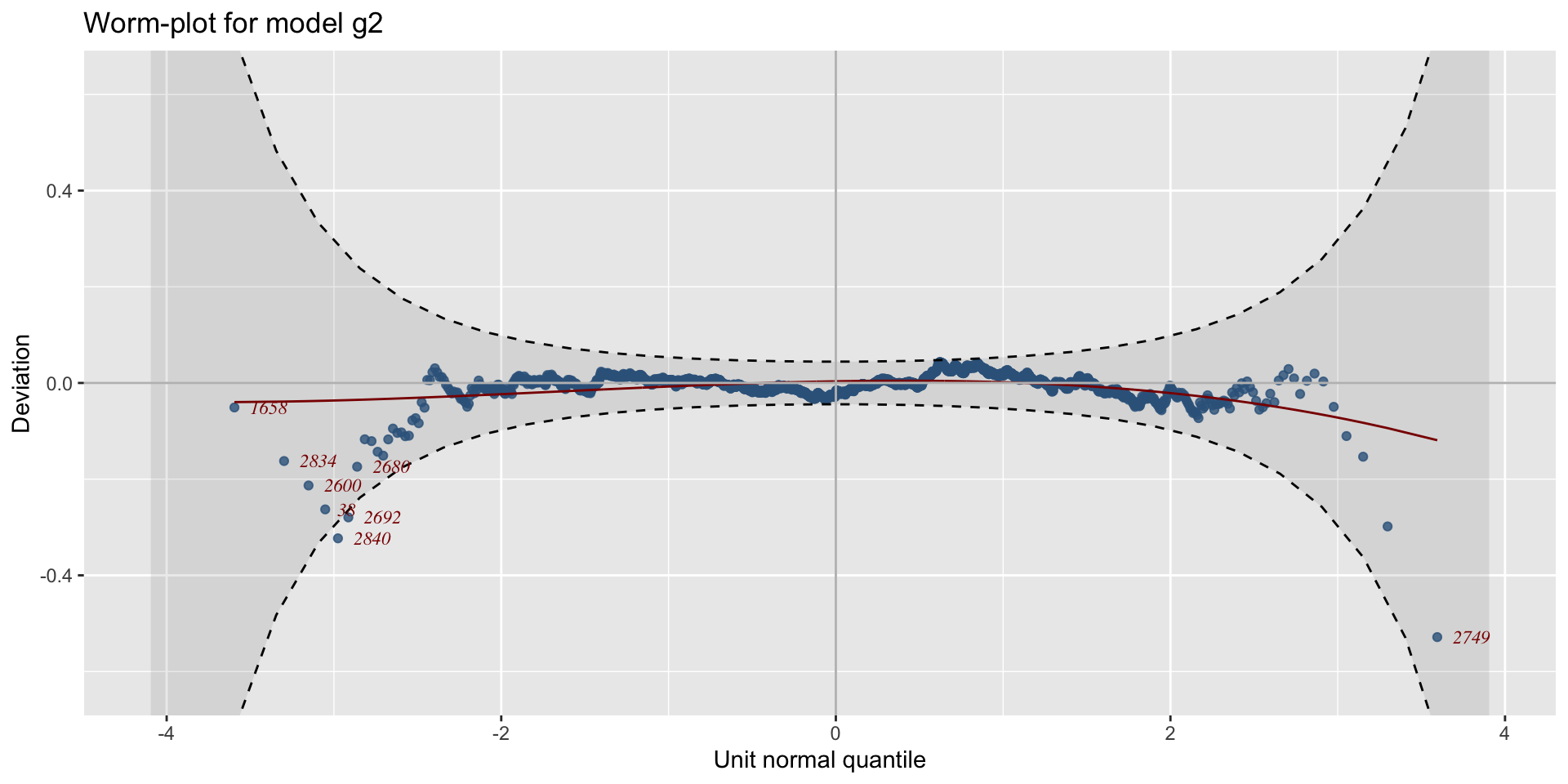
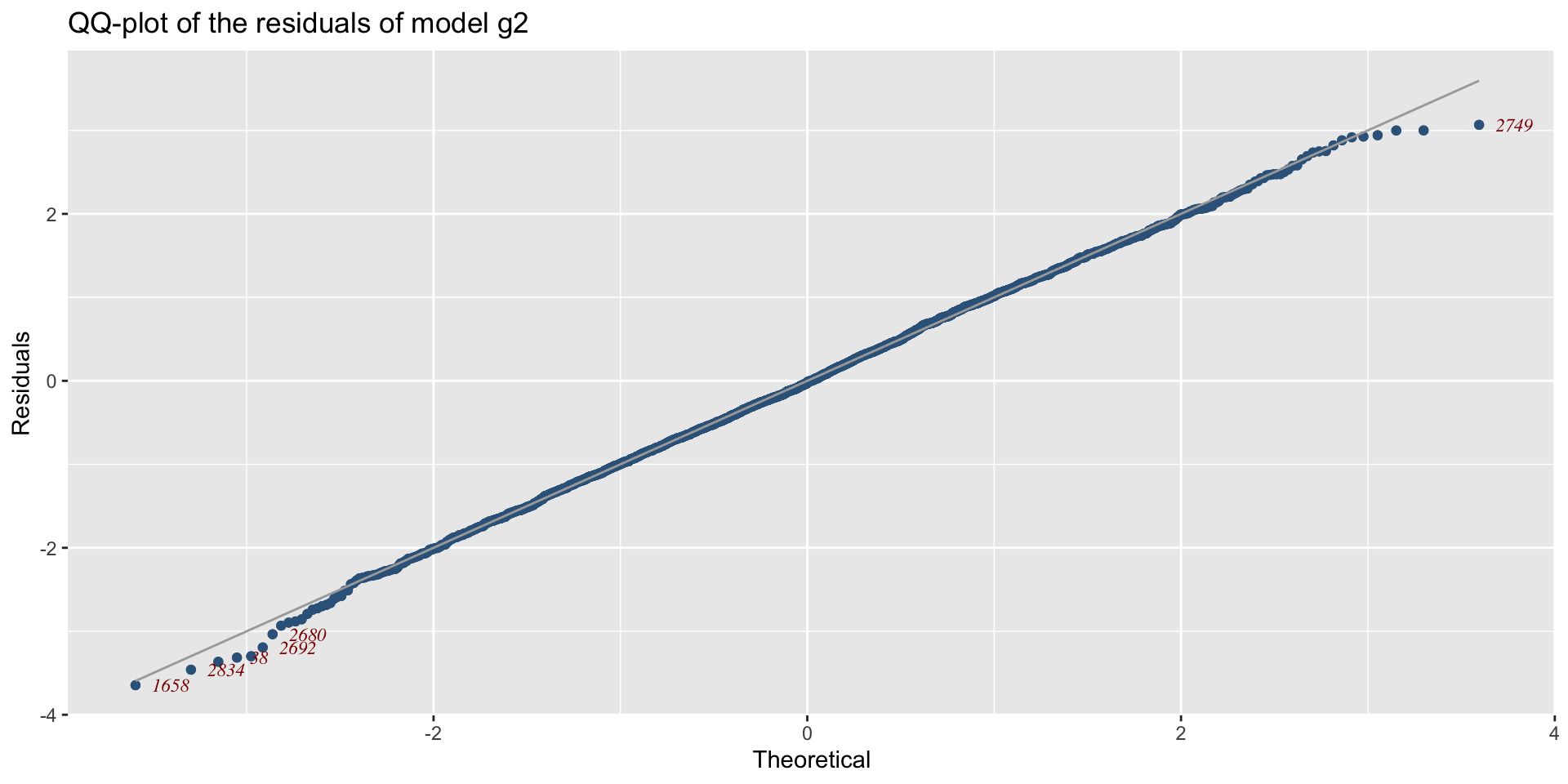
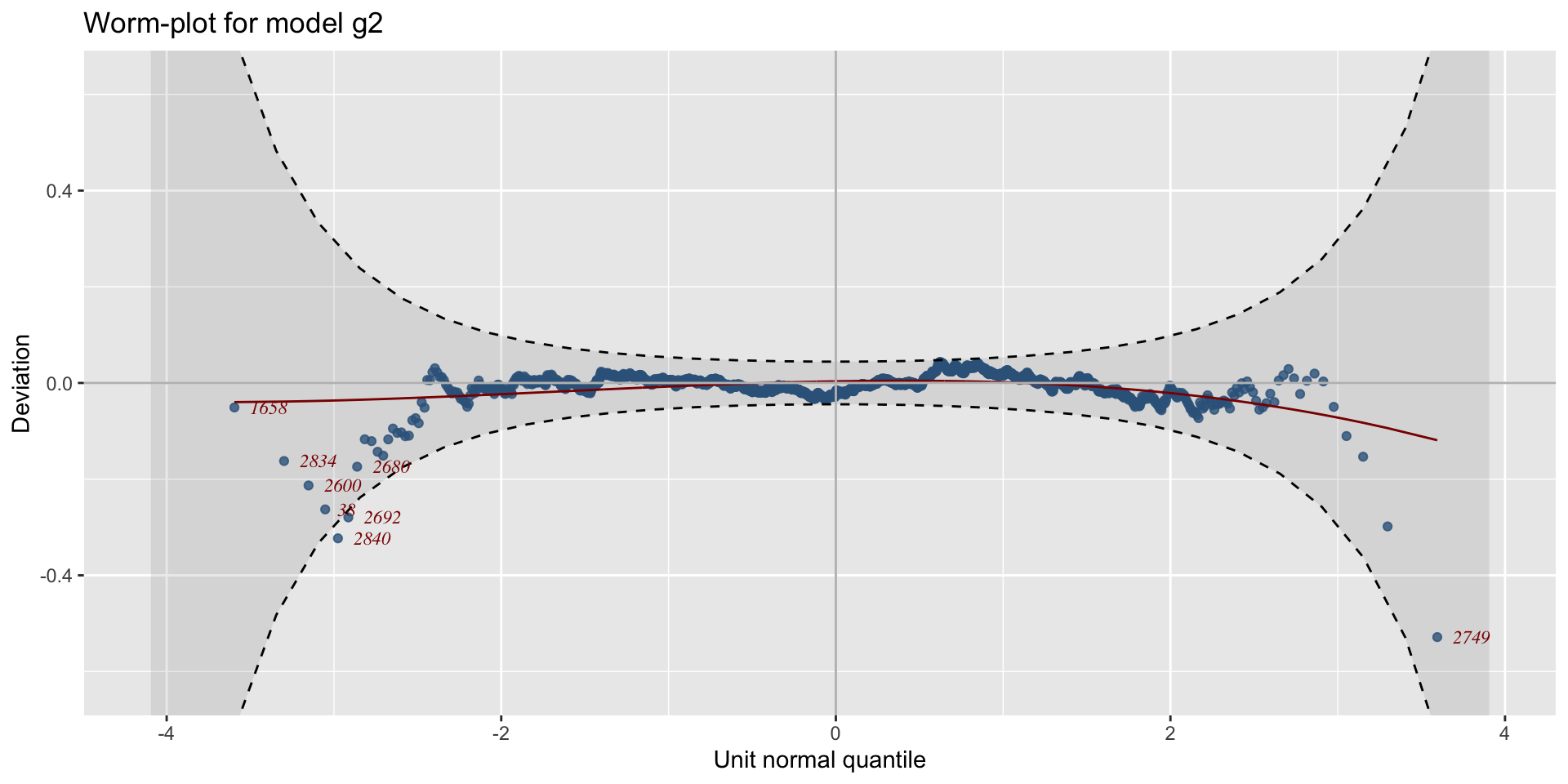
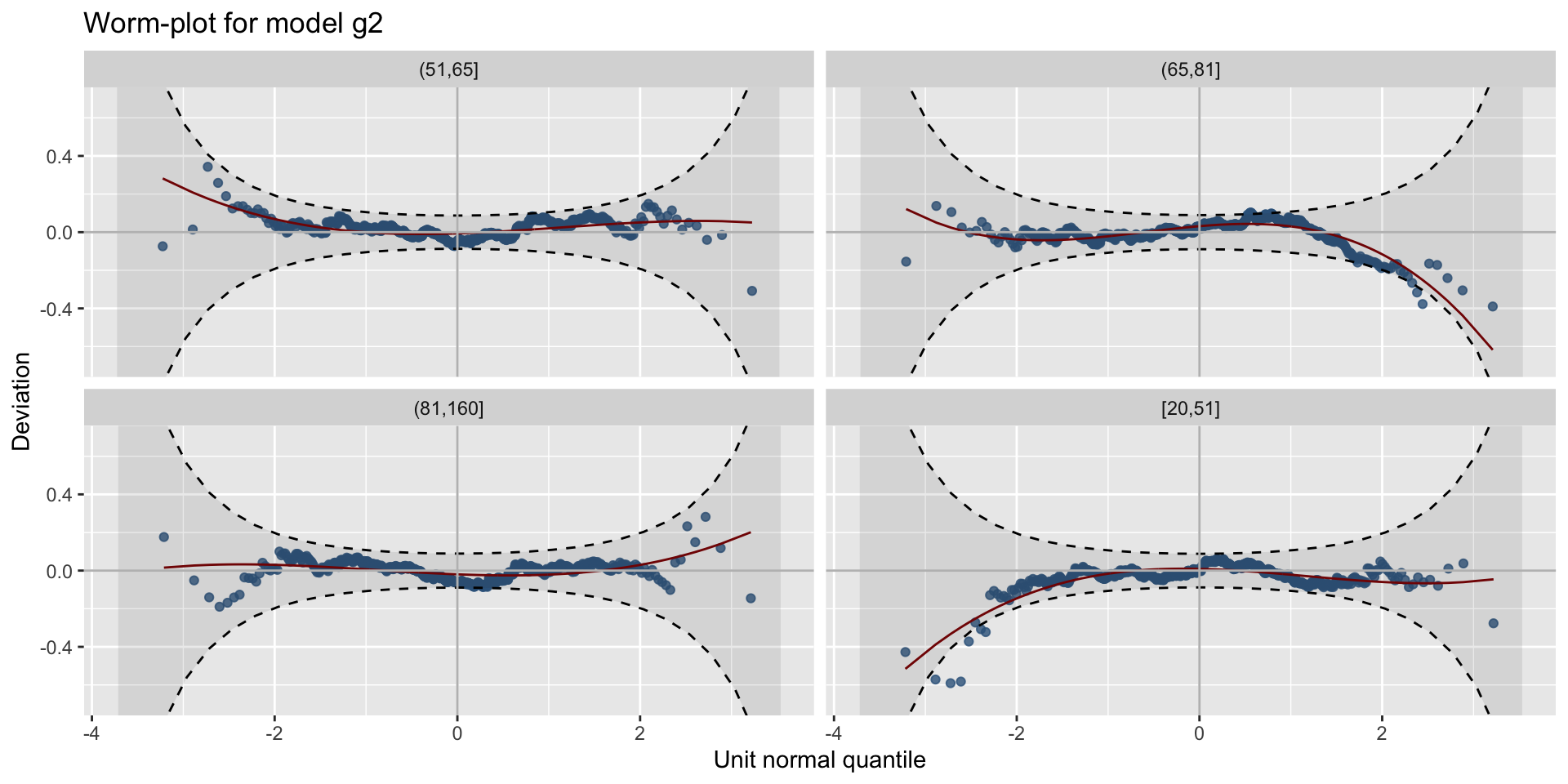
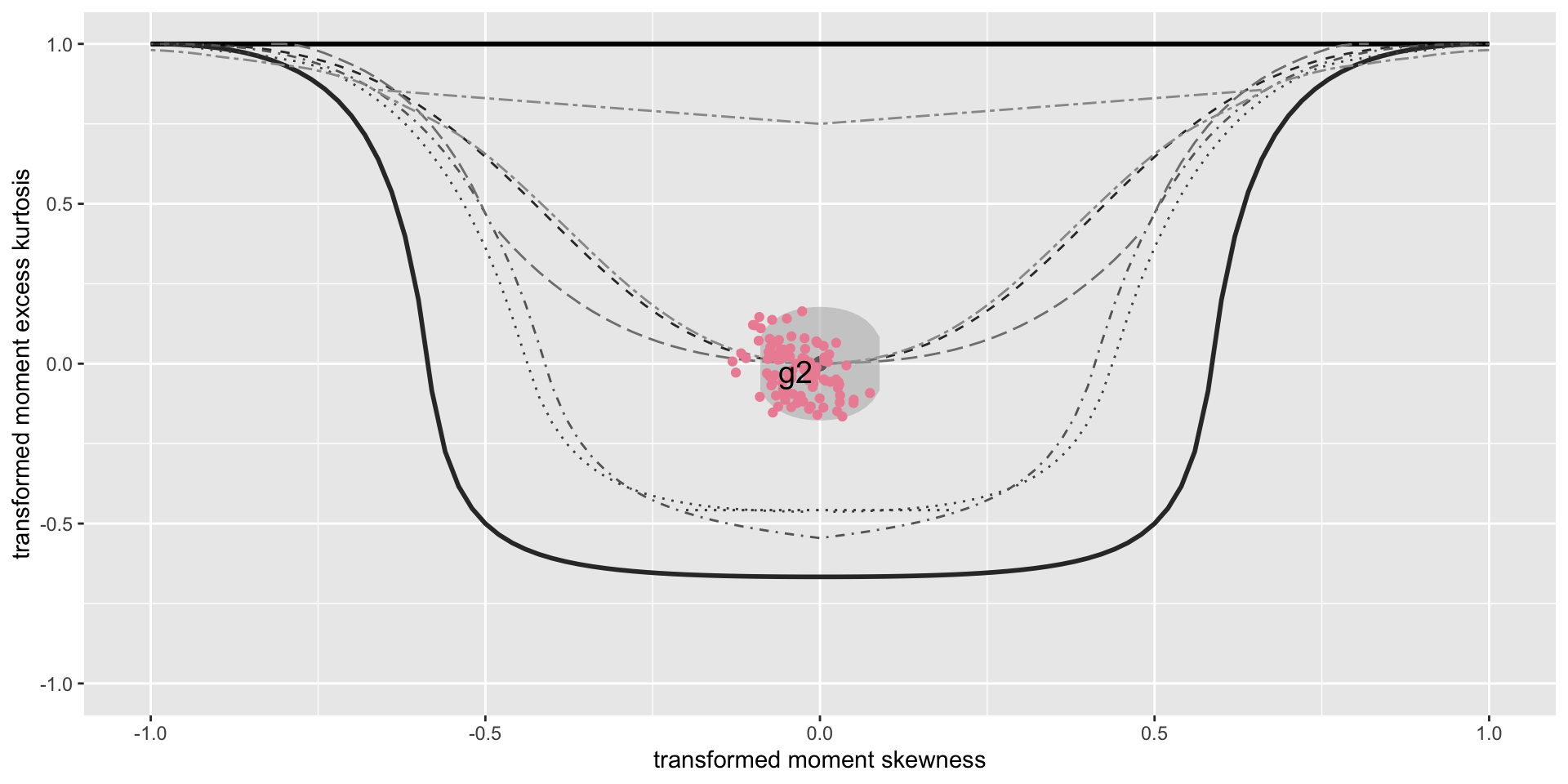
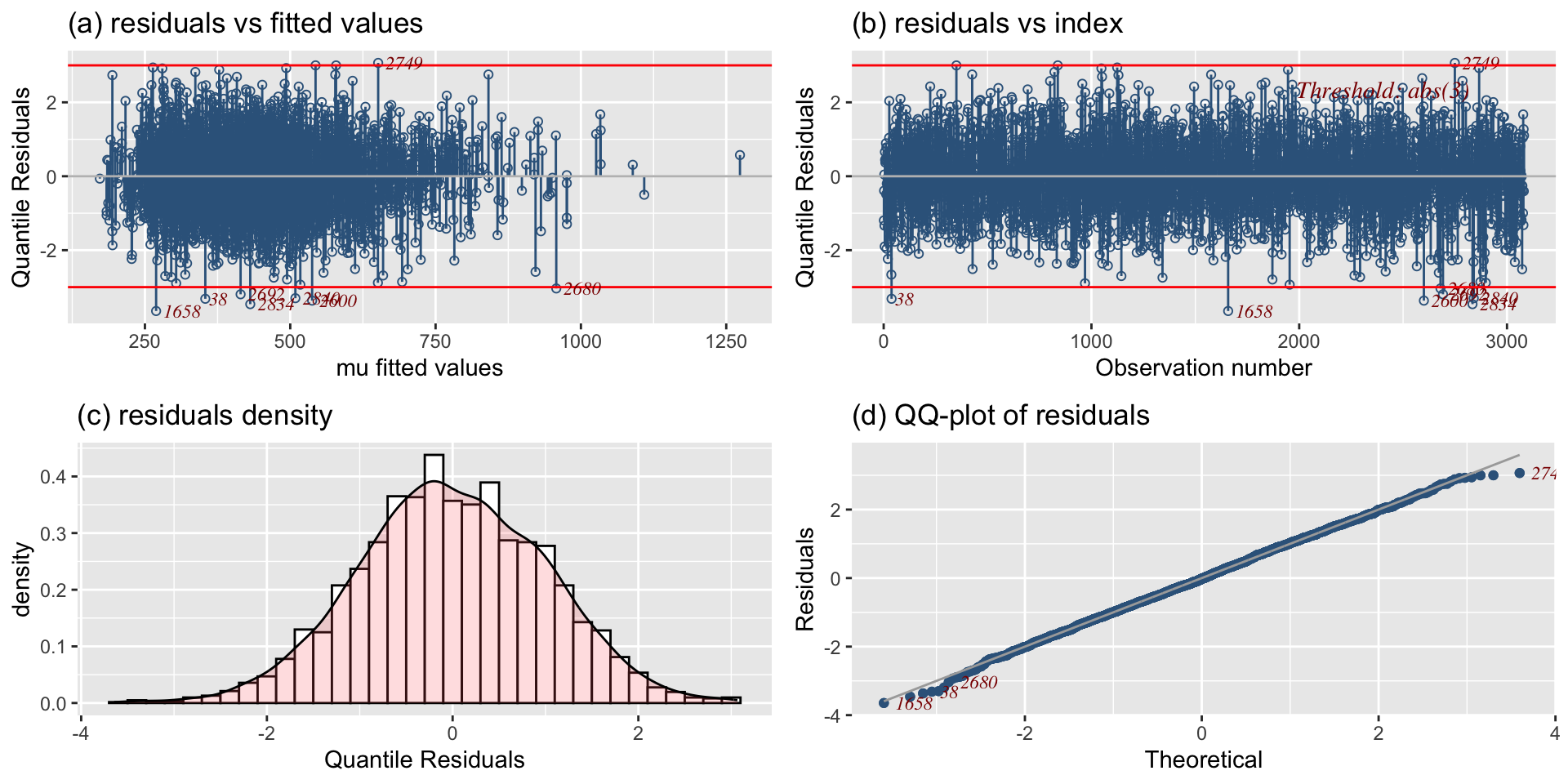
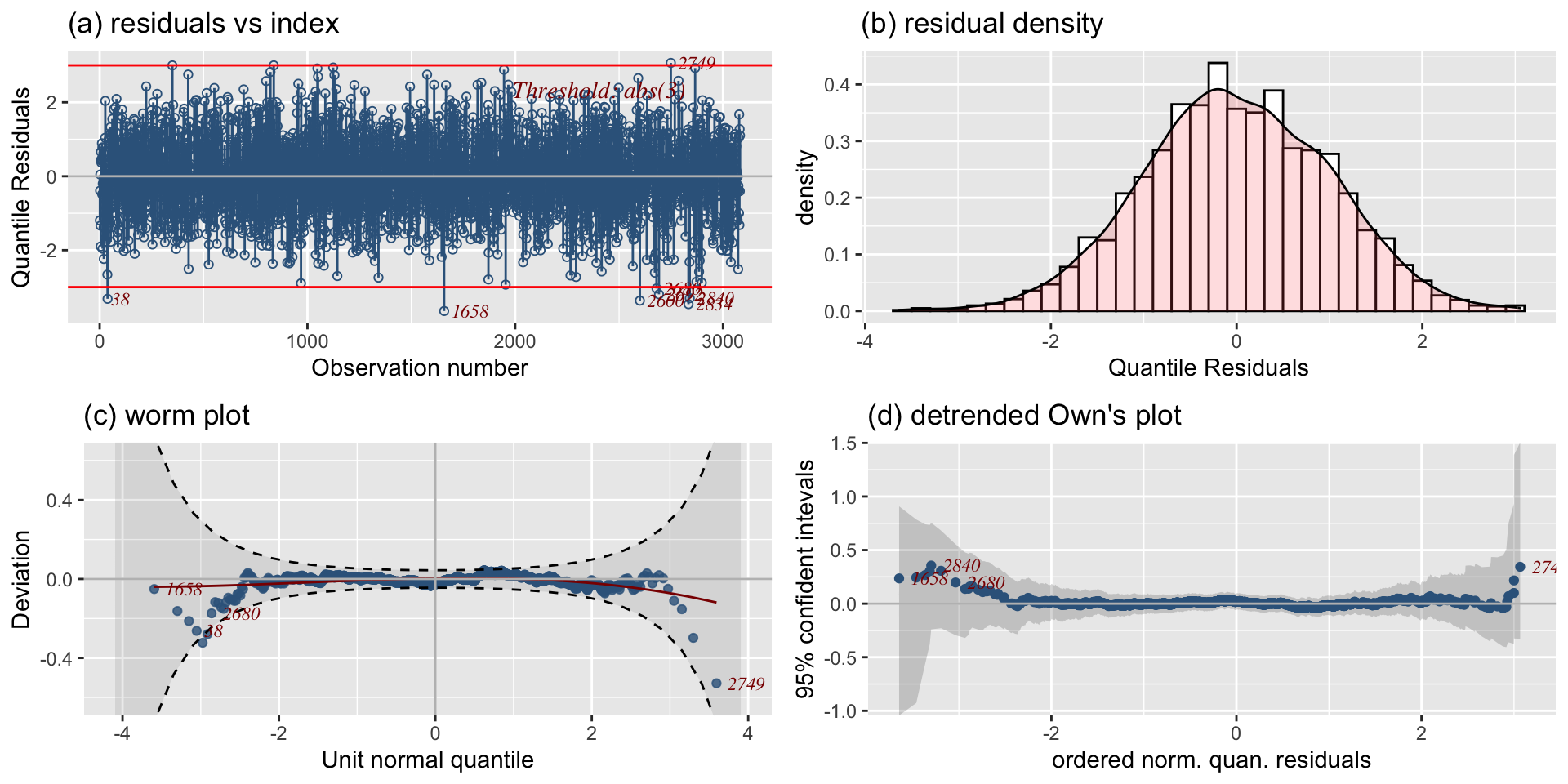
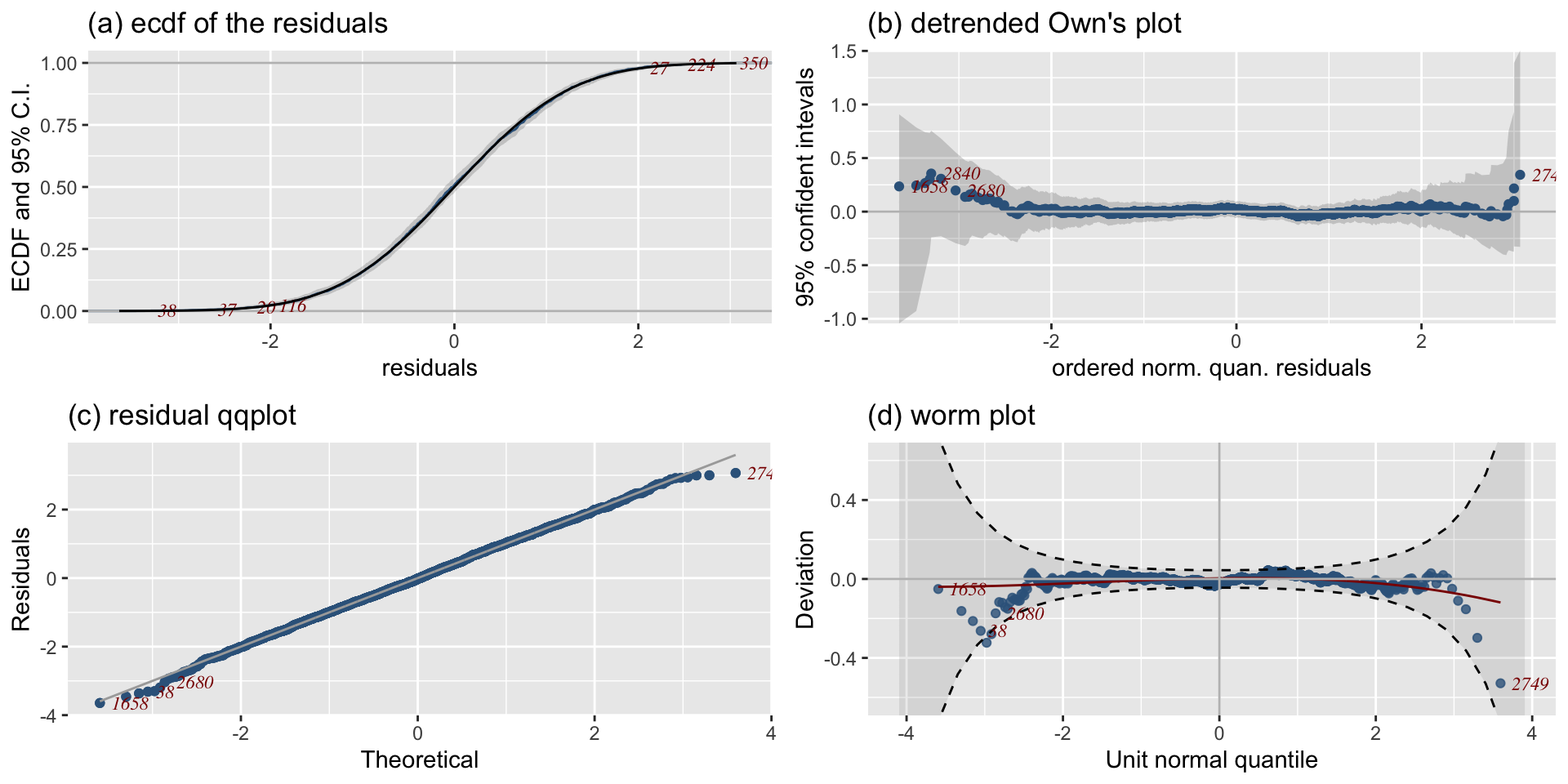
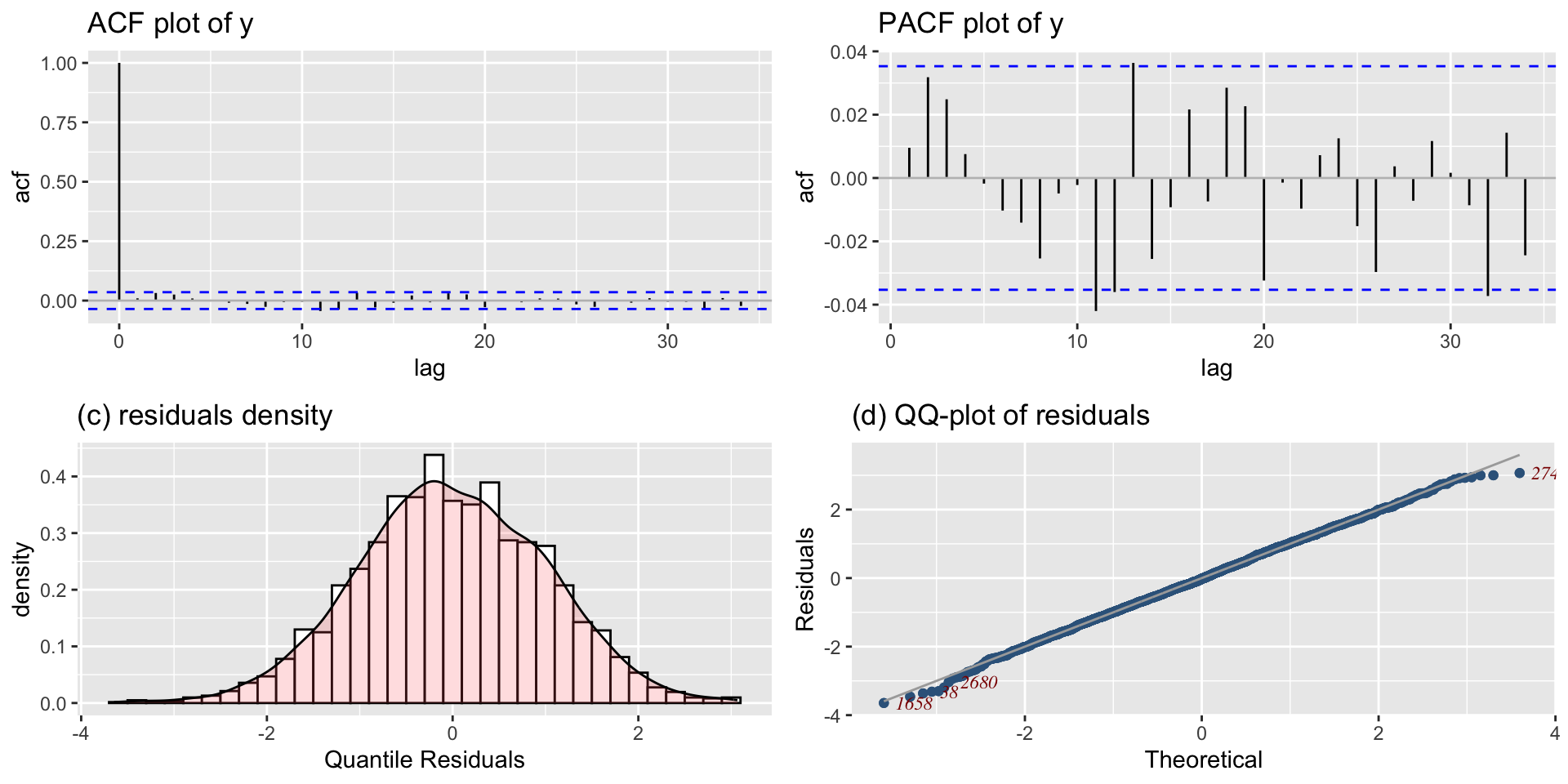
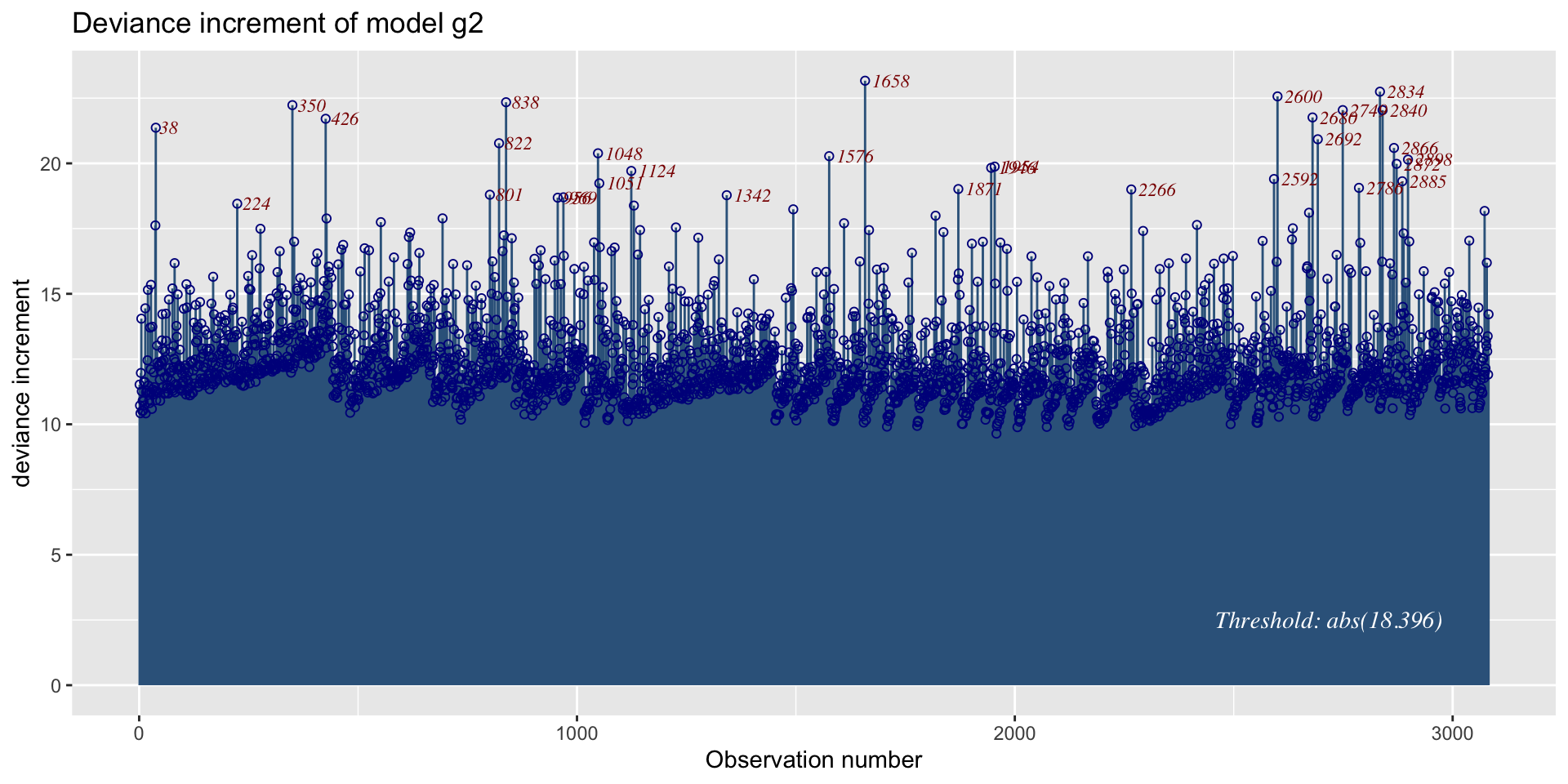
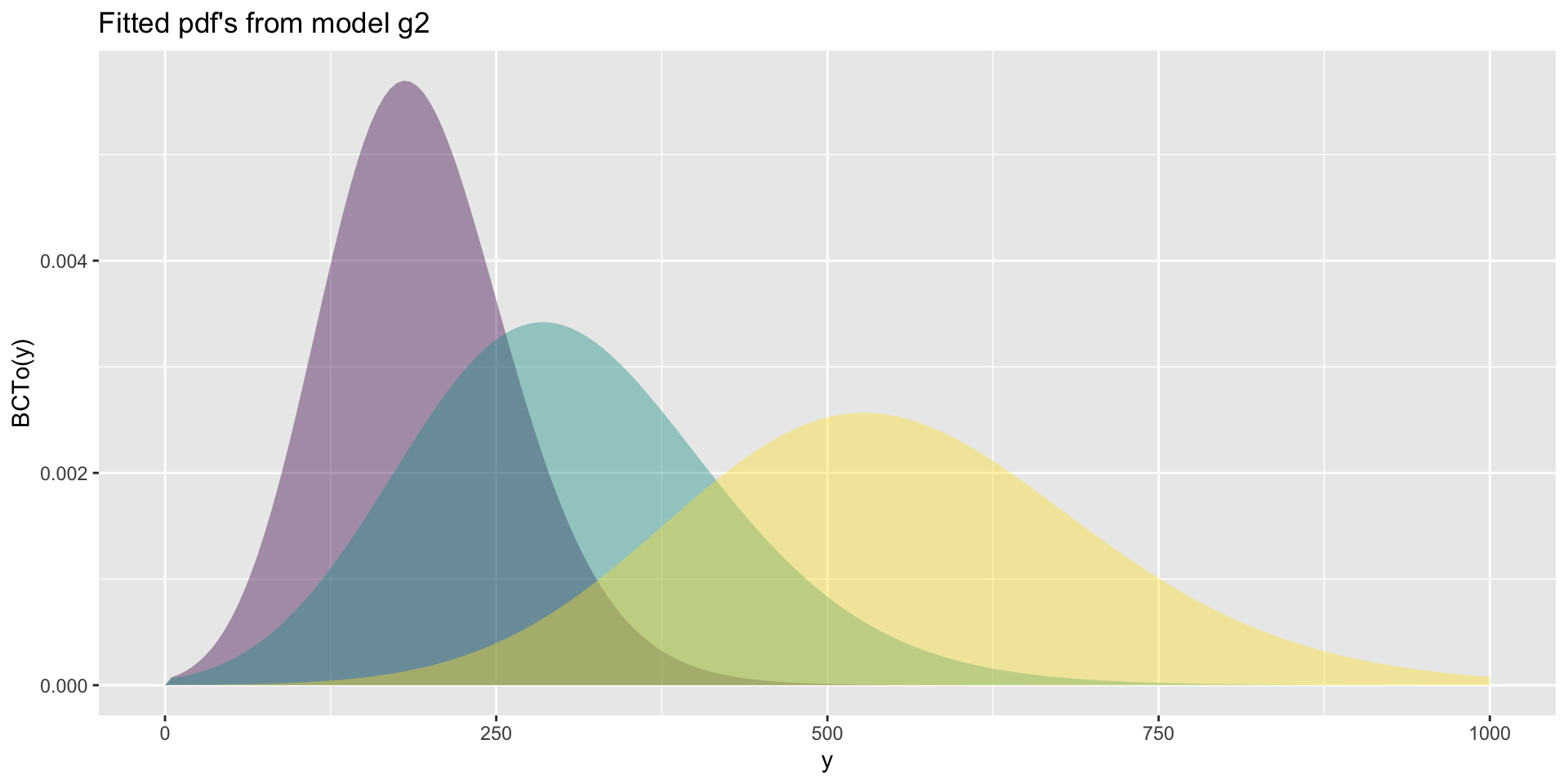
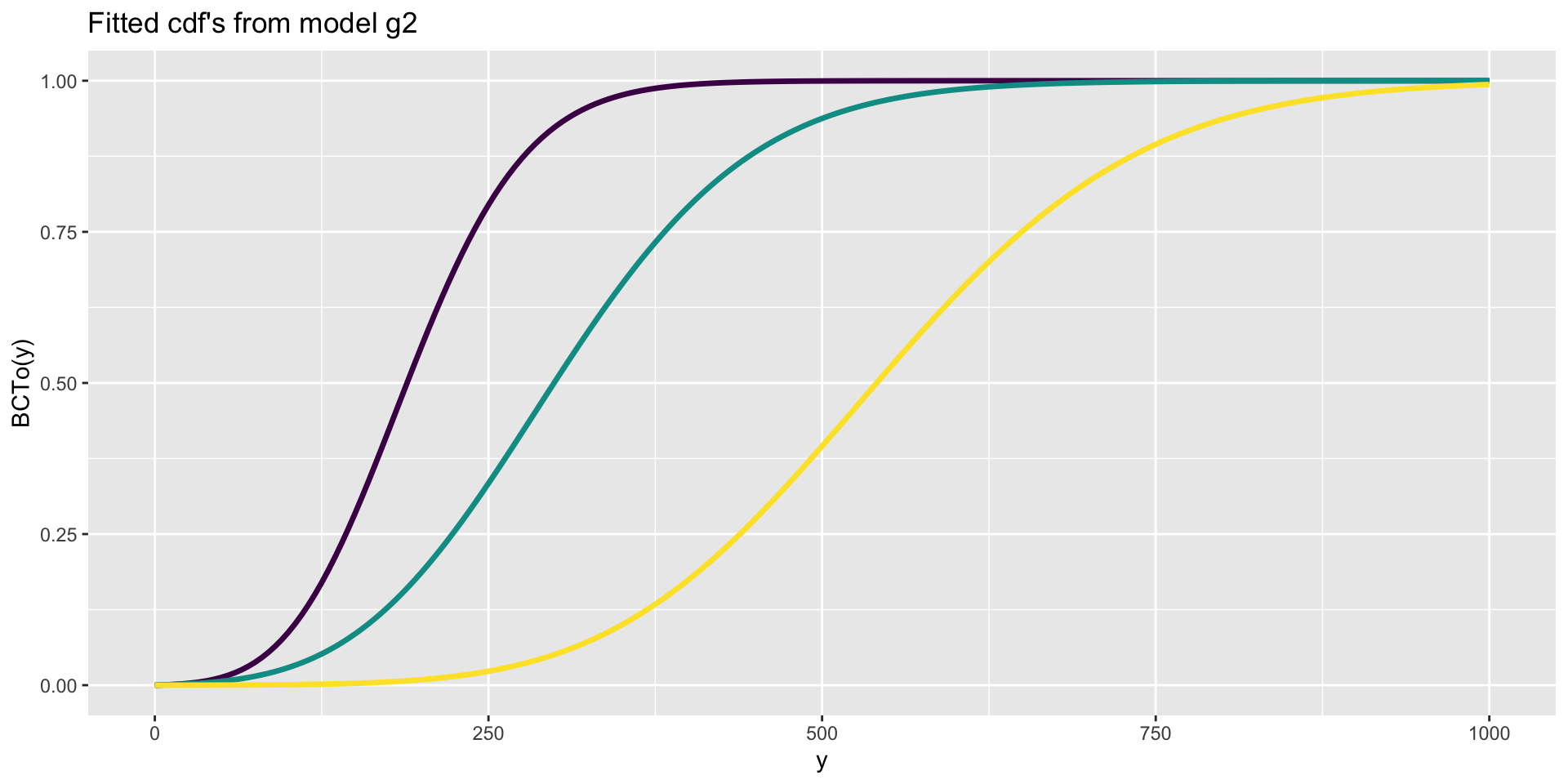
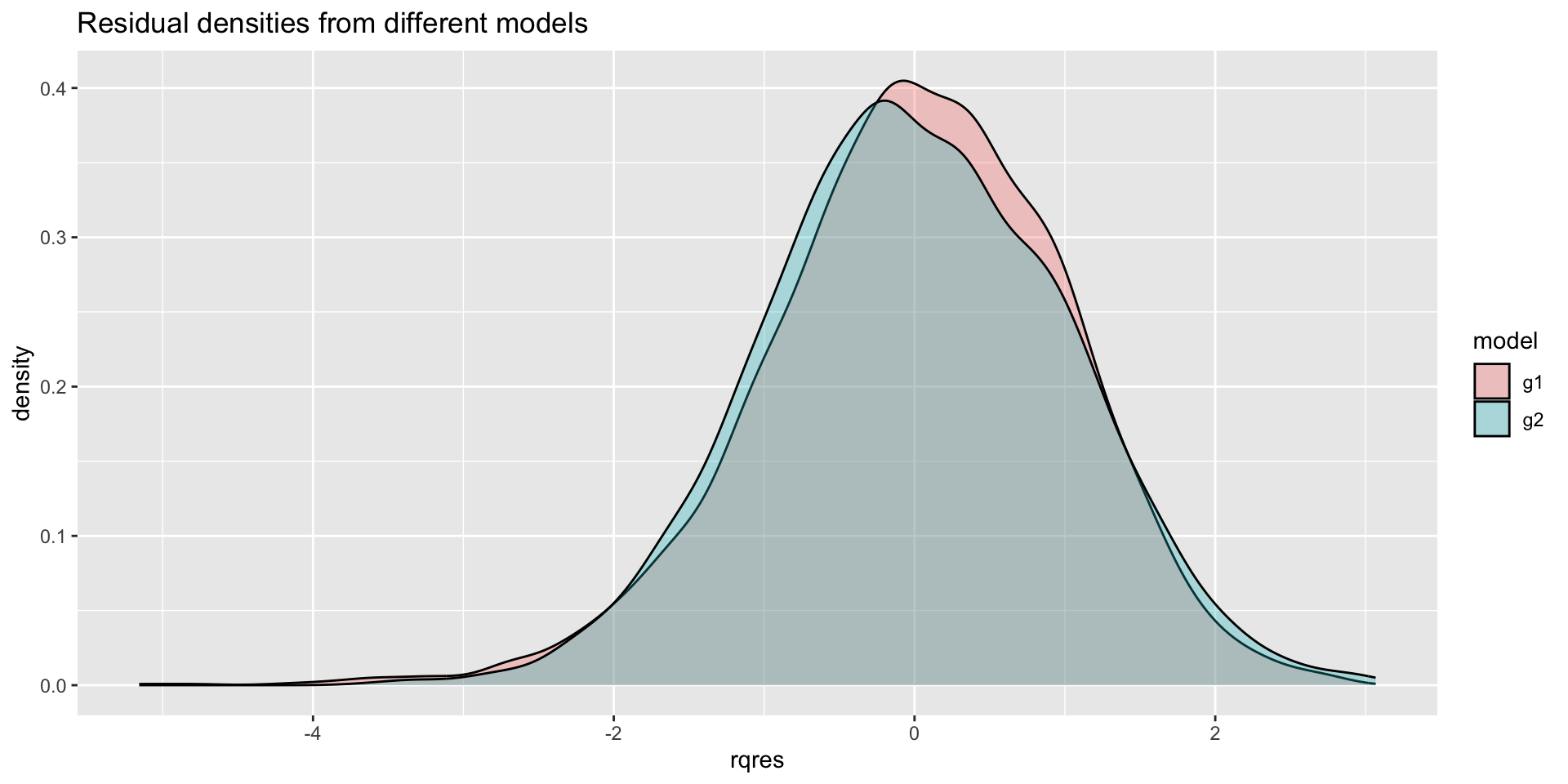
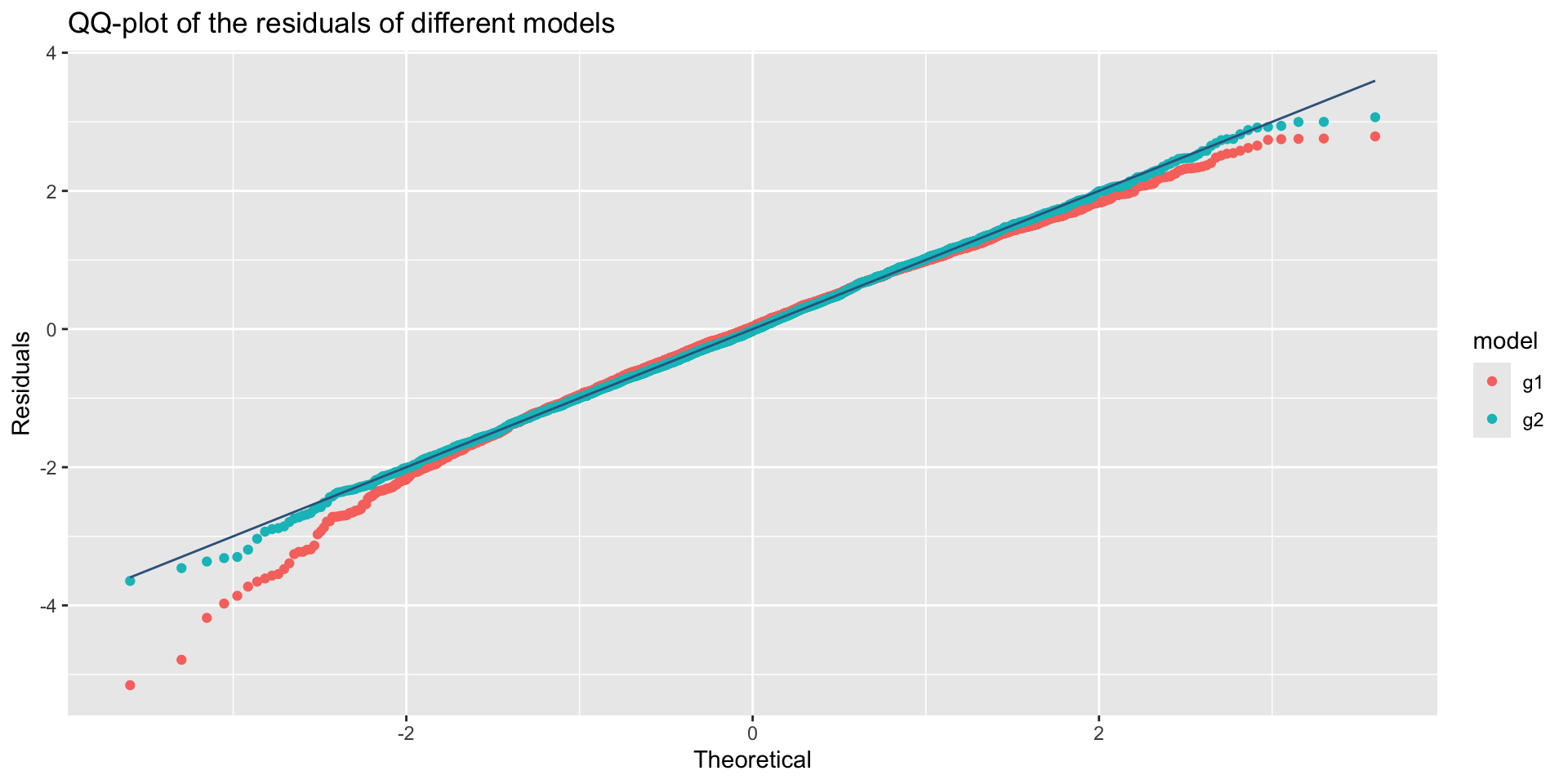
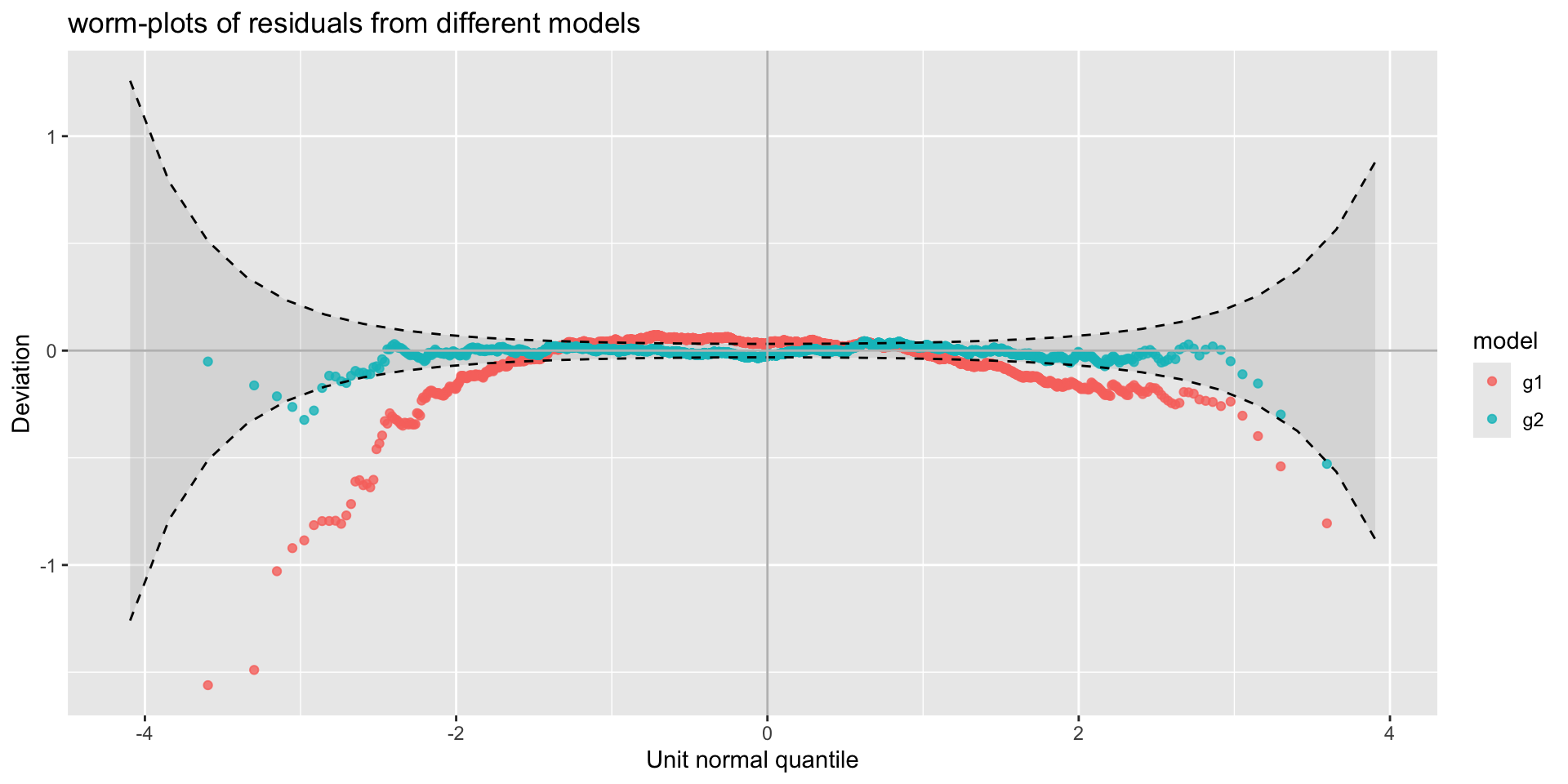
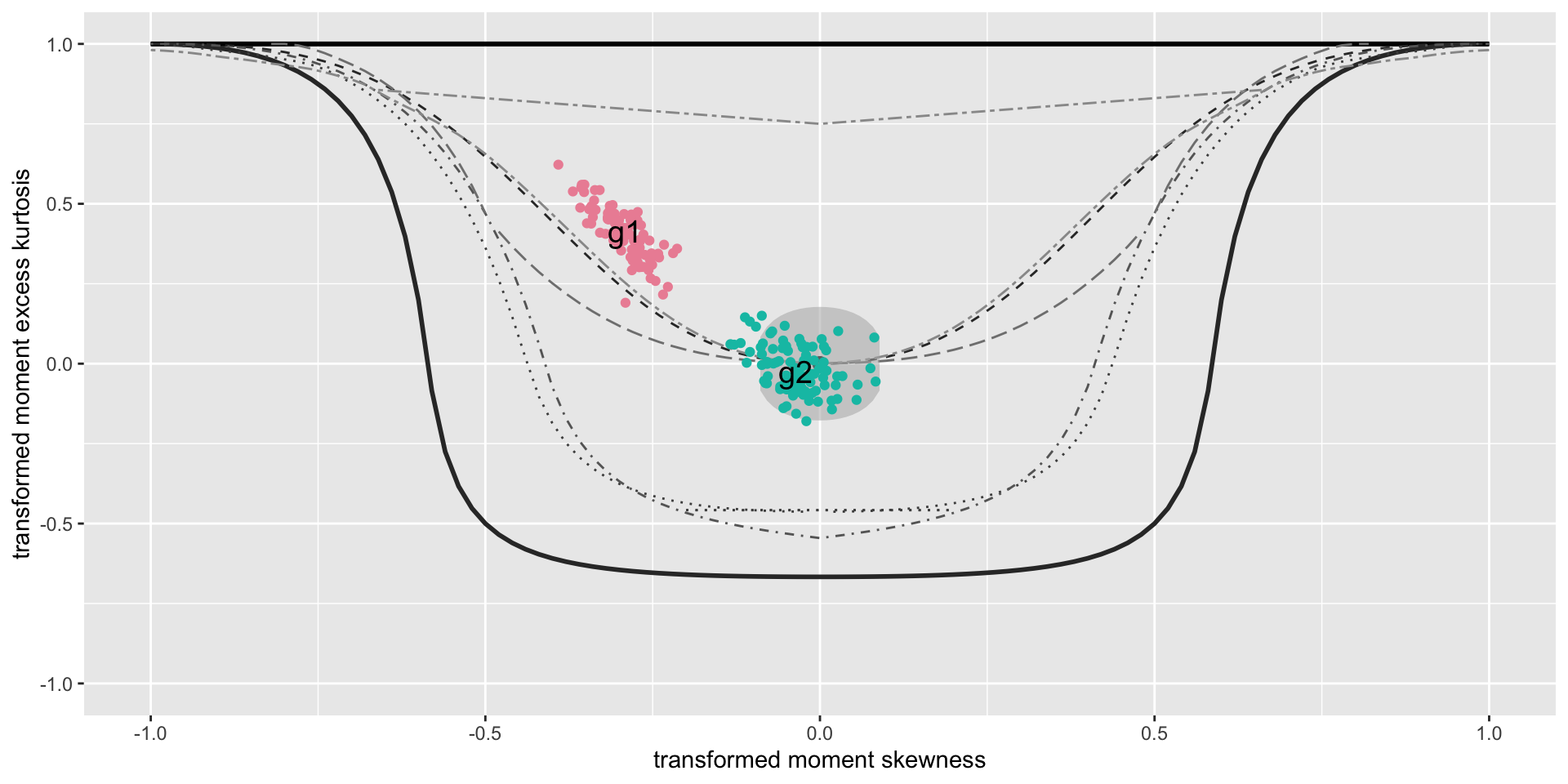
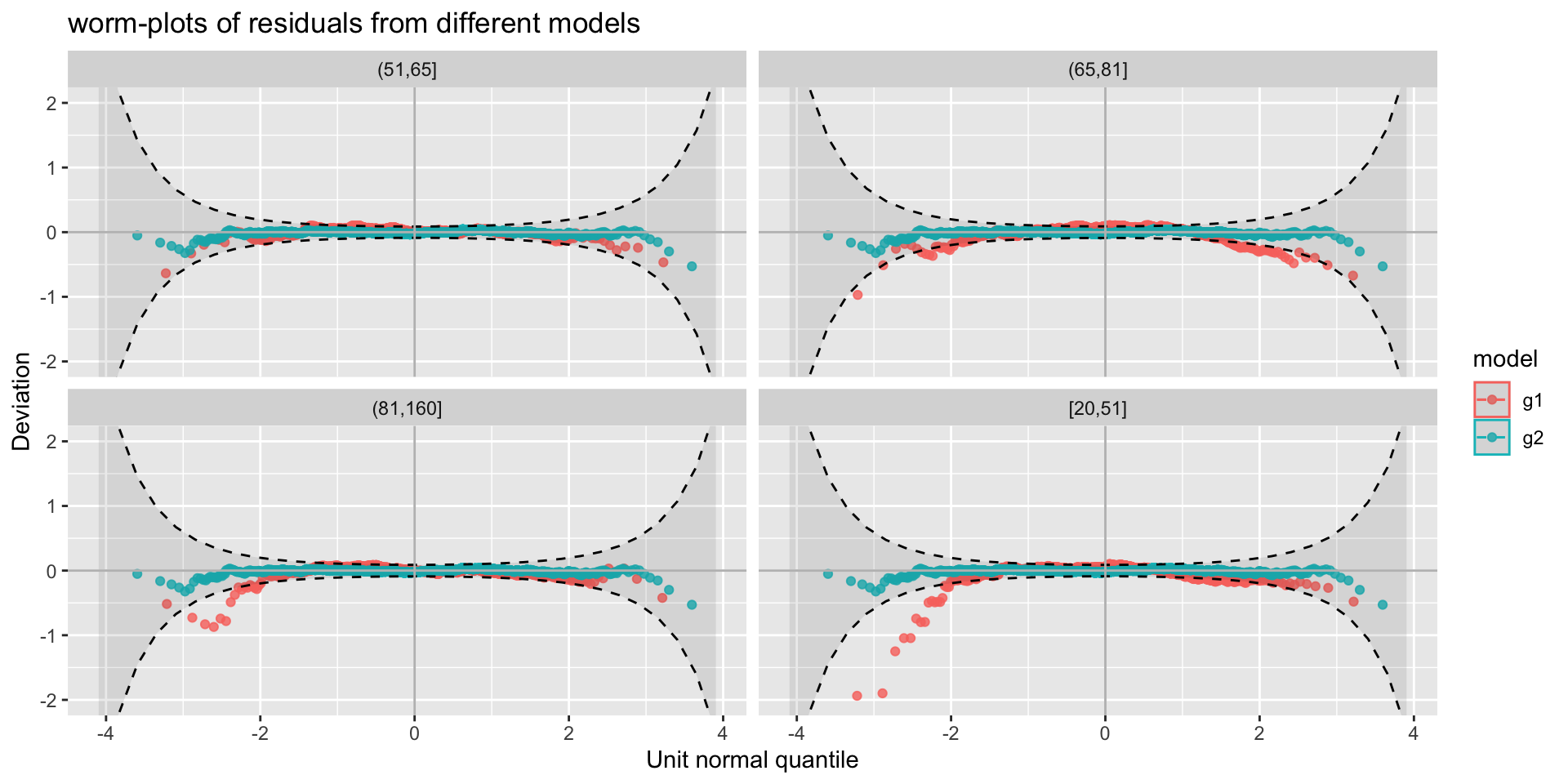


 The Books
The Books