Comparing Models
Mikis Stasinopoulos
Bob Rigby
Fernanda De Bastiani
Gillian Heller
Niki Umlauf
Introduction
what to compare
- distributions
- x-variables
how ro compare
graphicaldiagnostic tools;- model summary
statistics
summary statistics
| Data | Methods |
|---|---|
all data |
\(\chi^2\), GAIC |
test data |
LS, CRPS, MSE etc. |
data partitioning |
|
| K-fold | LS, CRPS |
| bootstrap | LS, CRPS |
Summary Comparison Statistics
- if
no partitionevaluation is done in the training dataset- Generalized AIC
\[ GAIC = \hat{GD}+ k \times df \] - \(\chi^2\) test:
likelihood ratiotest for nested models \[LR= GD_1/ GD_0 \sim \chi^2(df0- df_1) \]
- Generalized AIC
Summary Comparison Statistics (con.)
partition: evaluation is done onnewdata- Likelihod score (
LS) \(\equiv\) Prediction Global Deviance (PGD) - Continuous Rank Probabily Score (
CRPS) - Mean Absolute Prediction Error (
MAPE) - e.t.c.
- Likelihod score (
no partition
GAIC
| models | df | AIC | \(\chi^2\) | BIC |
|---|---|---|---|---|
| mlinear | 24 | 22808.89 | 22855.45 | 22953.69 |
| madditive | 29.24 | 22814.33 | 22871.07 | 22990.77 |
| mfneural | 160 | 22930.36 | 23240.76 | 23895.69 |
| mregtree | 30 | 38754.64 | 38812.84 | 38935.64 |
| mcf | 404 | 38754.64 | 24481.28 | 26134.99 |
GAIC (continuous)
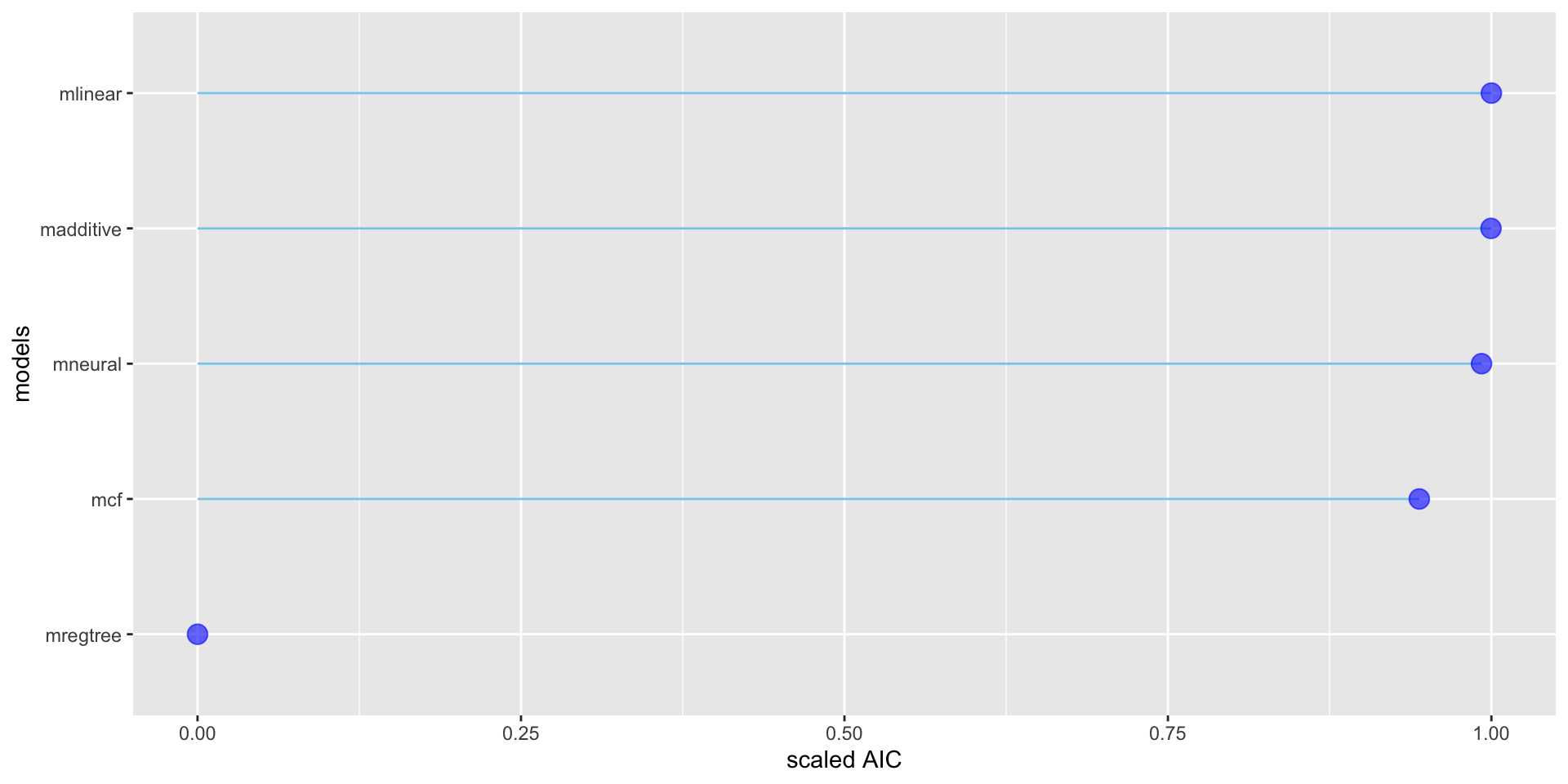
Figure 1: A lollipop plot of AIC of the fitted models.
partition
prediction measures
- \[LS= \sum_{i=1}^{n^*} \log \left[y^*_i | \hat{\theta}_i \left(\textbf{x}_i^*\right) \right] \]
- \[CRPS = -\sum_{i=1}^{n} \int \left(F(y| \hat{\theta}_i \left(\textbf{x}_i^*\right) -\textbf{I}\left(y \ge y^*_i\right)\right)^2 dy,\]
- \[MAPE= \texttt{med} \left(\left|100 \left(\frac{\hat{\mu}_i(\textbf{x}_i^*)-y^*}{y^*}\right) \right|_{i=1,\ldots.n}\right)\]
prediction measures table
| models | LS | CRPS | |
|---|---|---|---|
| mlinear | -6.2302 | 73.5738 | |
| madditive | -6.2288 | 73.8201 | |
| mfneural | -6.5126 | 79.9394 | |
| mregtree | -6.2930 | 78.6704 | |
| mcf | -6.2966 | 79.0644 |
summary
the GAIC is well established and tested (the df of freedom need to be known)
the linear and additive model are good when there are not many explanatory variables (but somehow interactions have to be considered)
more work has to be done to standardised all ML techniques so their partitioning of data are comparable to the conventional additive models
end


 The Books
The Books

www.gamlss.com