Continuous Distributions
Types
\((-\infty, \infty)\) real line \(\Re\) [Chapter 4 of Rigby et al (2019)]
\((0, \infty )\) positive real line \(\Re^{+}\) [Chapter 5]
\((0,1)\) real line on interval \(\Re_{(0,1)}\) (not containing zero or one) [Chapter 6]
Explicit distributions in real line, \(\Re\).
Location and Scale family
if \[Y\sim {D}(\mu,\sigma,\nu,\tau)\] then \[\varepsilon=(Y-\mu)/\sigma\sim {D}(0,1,\nu,\tau),\]
i.e. \(Y=\mu+\sigma\varepsilon\), so \(Y\) is a scaled and shifted version of the random variable \(\varepsilon\).
location
scale
2 parameter in \(\Re\)
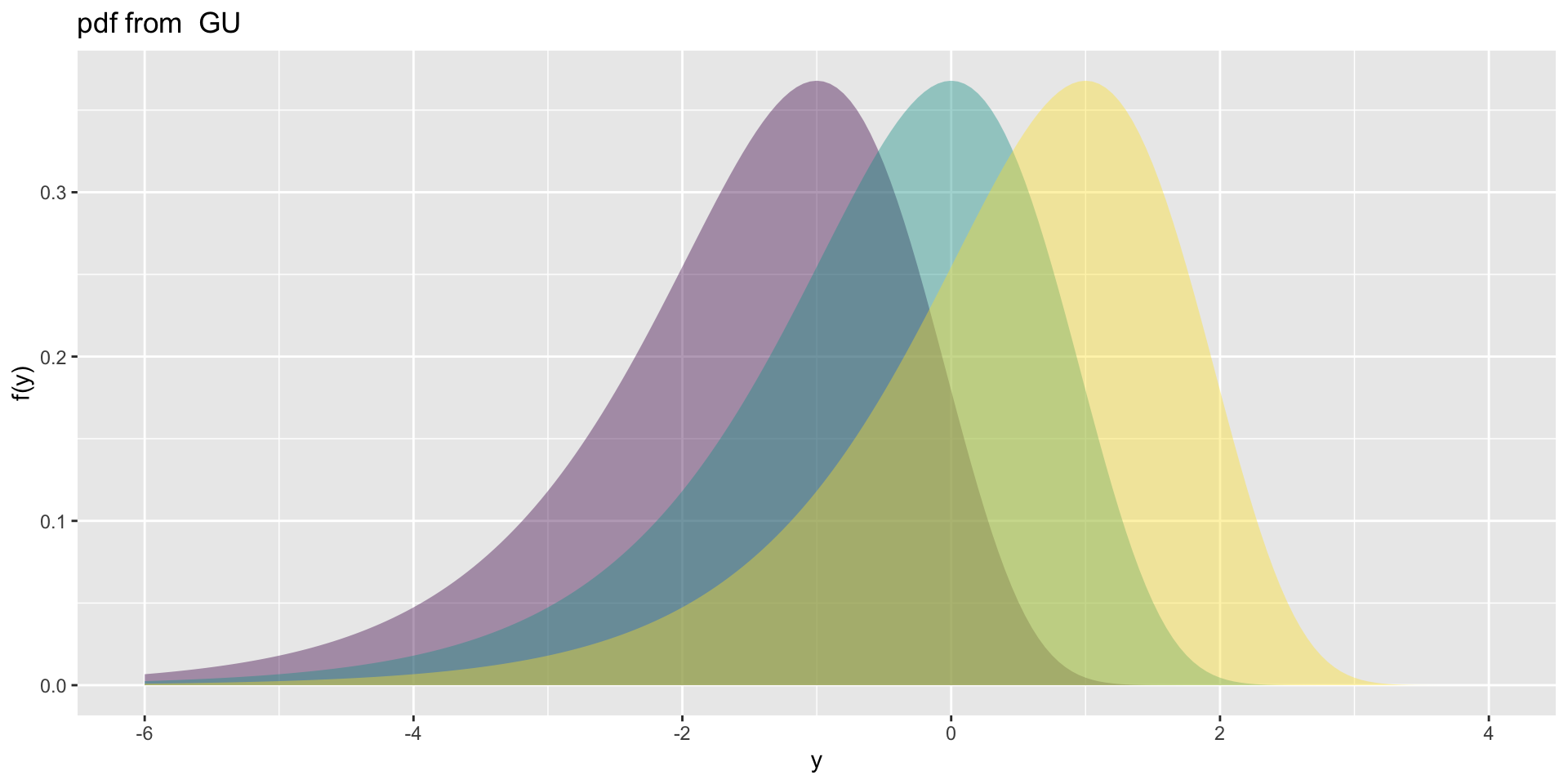
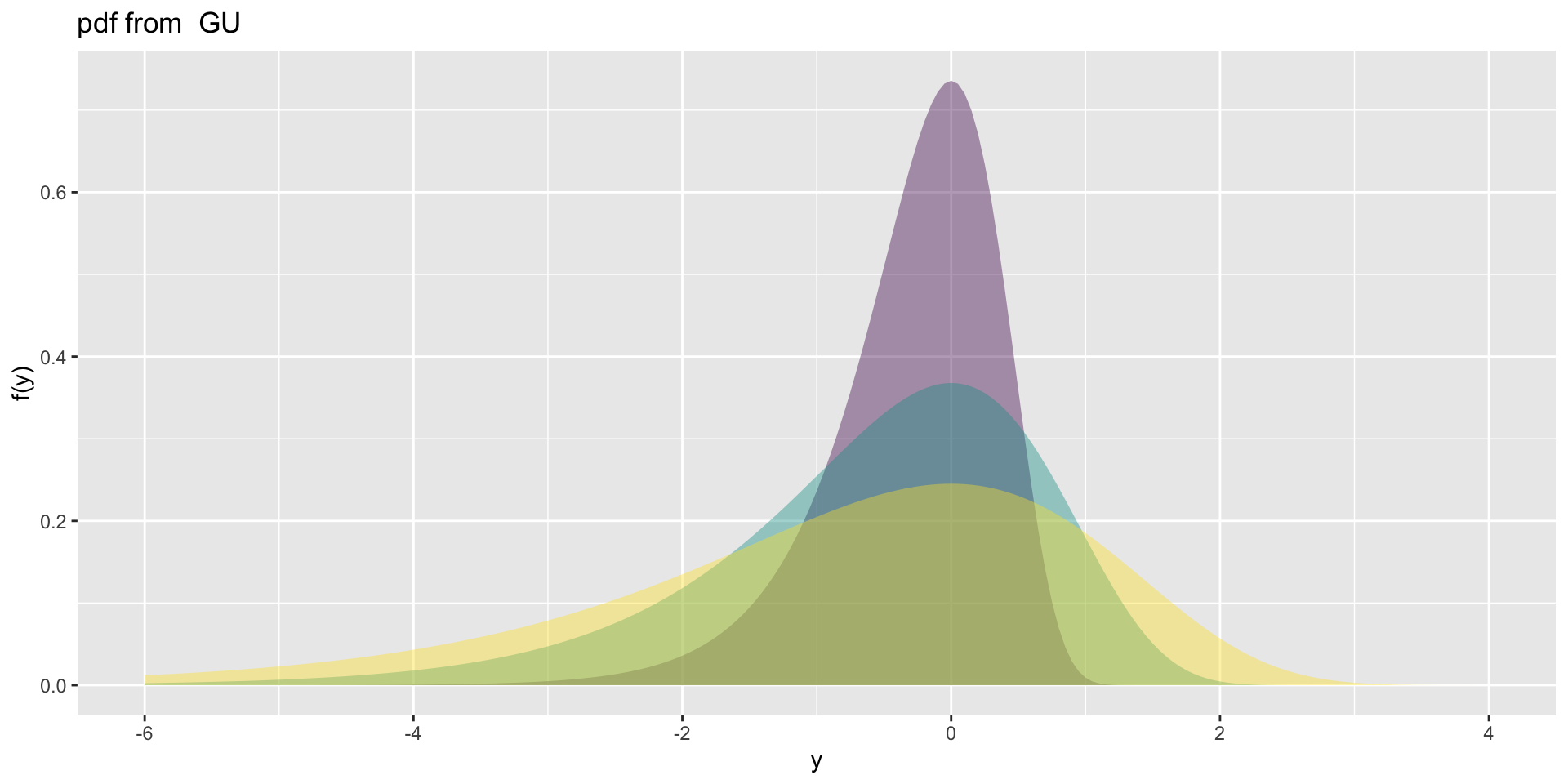
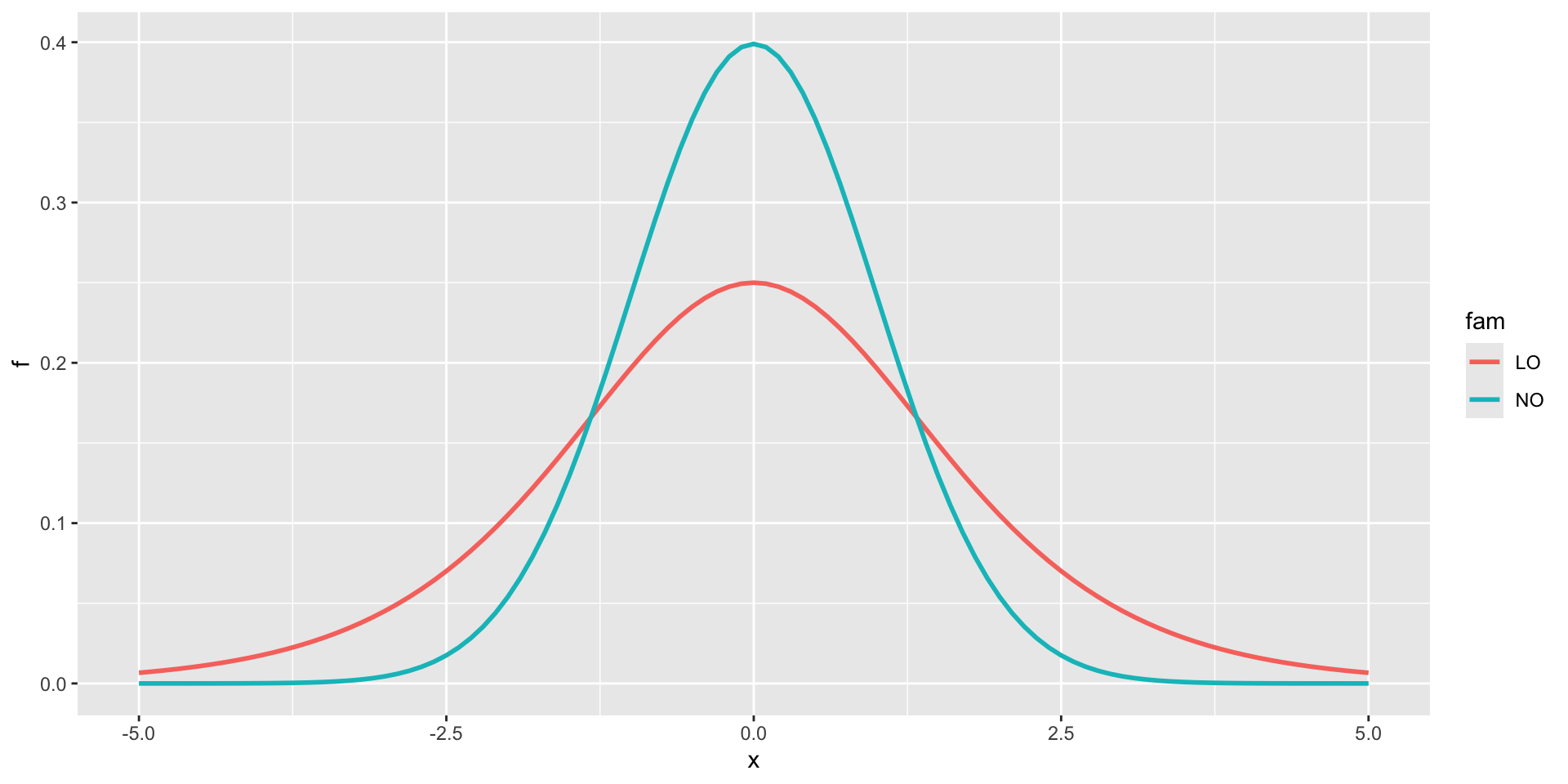
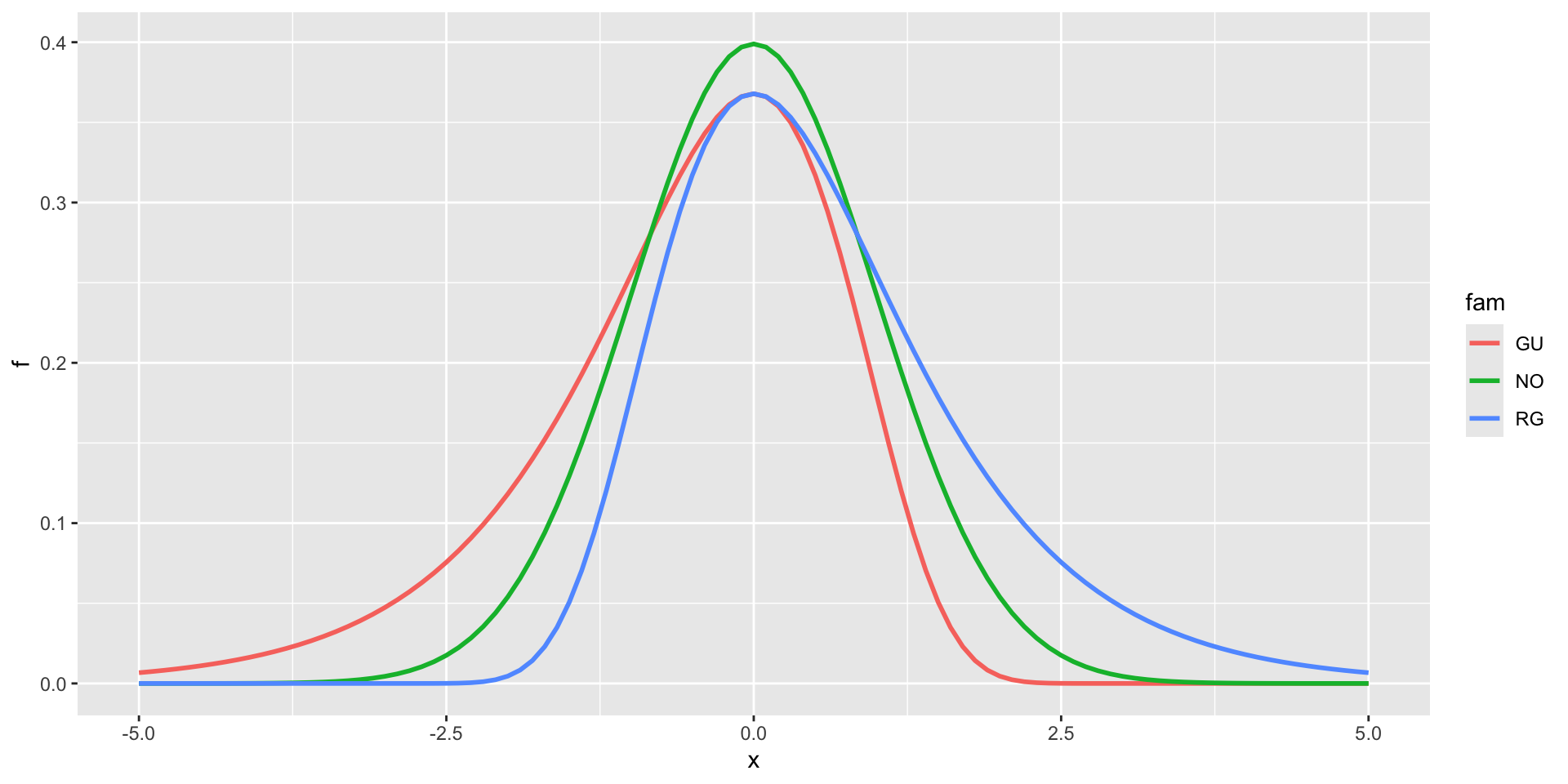
Gumbel, GU(\(\mu\), \(\sigma\)), left skewLogistic, LO(\(\mu\), \(\sigma\)), leptoNormal, NO(\(\mu\), \(\sigma\)), NO(\(\mu\), \(\sigma^2\))Reverse GumbelRG(\(\mu\), \(\sigma\)), right skew
2 parameter in \(\Re\) (con.)
NormalagainstLogistic
2 parameter in \(\Re\) (con.)
NormalagainstGumbelandreverse Gumbel
3 parameter in \(\Re\)
exponential Gaussian: exGAUS\((\mu, \sigma, \nu)\) for modelling right skew data,normal family:NOF\((\mu, \sigma, \nu)\) for modelling mean and variance relationships following the power law;power exponential:PE\((\mu, \sigma, \nu)\) and PE2\((\mu, \sigma, \nu)\) for modelling lepto and platy kurtotic data;t family: TF\((\mu, \sigma, \nu)\) and TF2\((\mu, \sigma, \nu)\) for modelling lepro kurtotic data;skew normal: SN1\((\mu, \sigma, \nu)\) and SK2\((\mu, \sigma, \nu)\) for modellinng skewness in data.
3 parameter in \(\Re\) (con.)
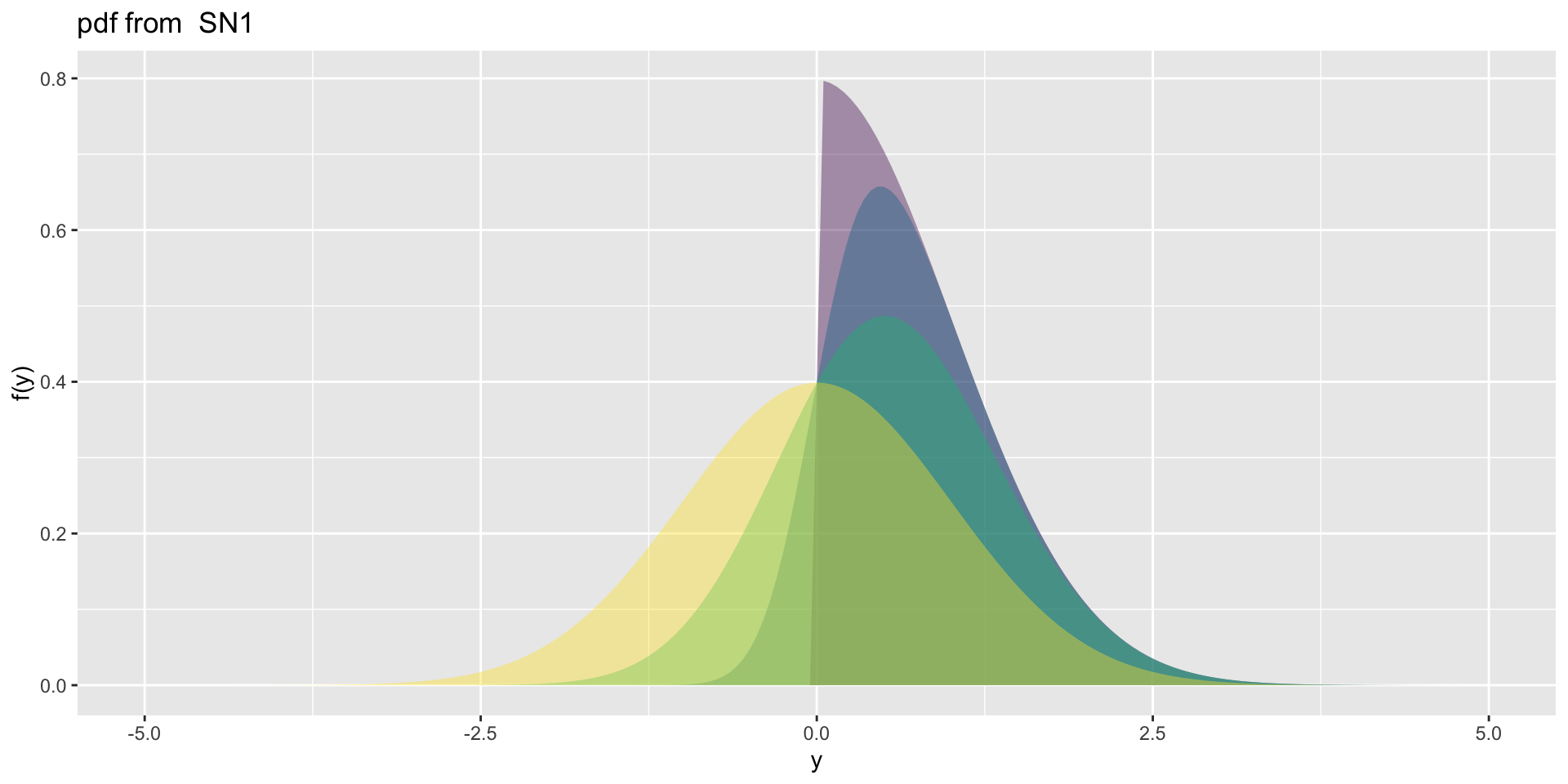
Skew Normal type 1
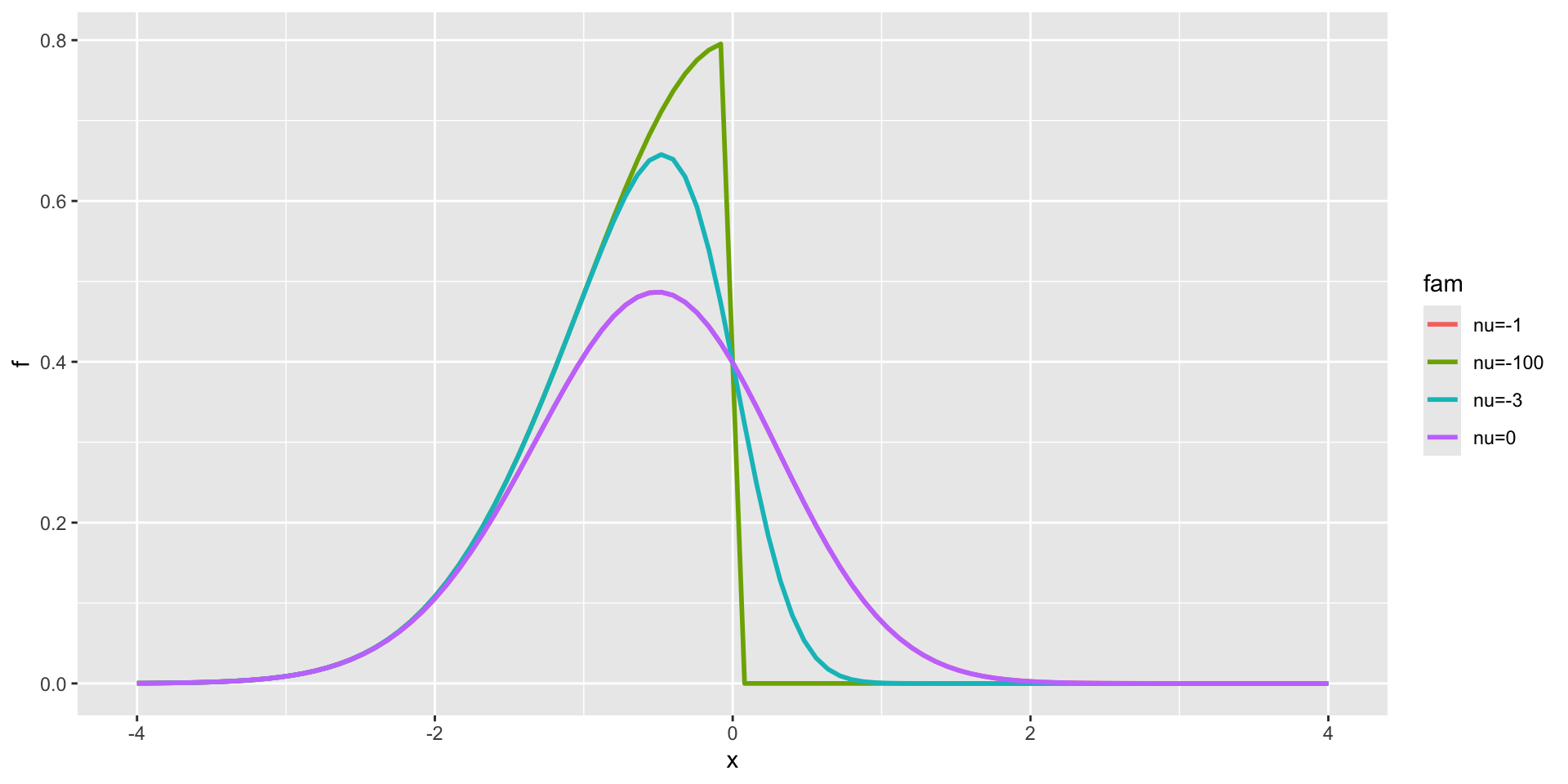
3 parameter in \(\Re\) (con.)
Skew Normal type 1
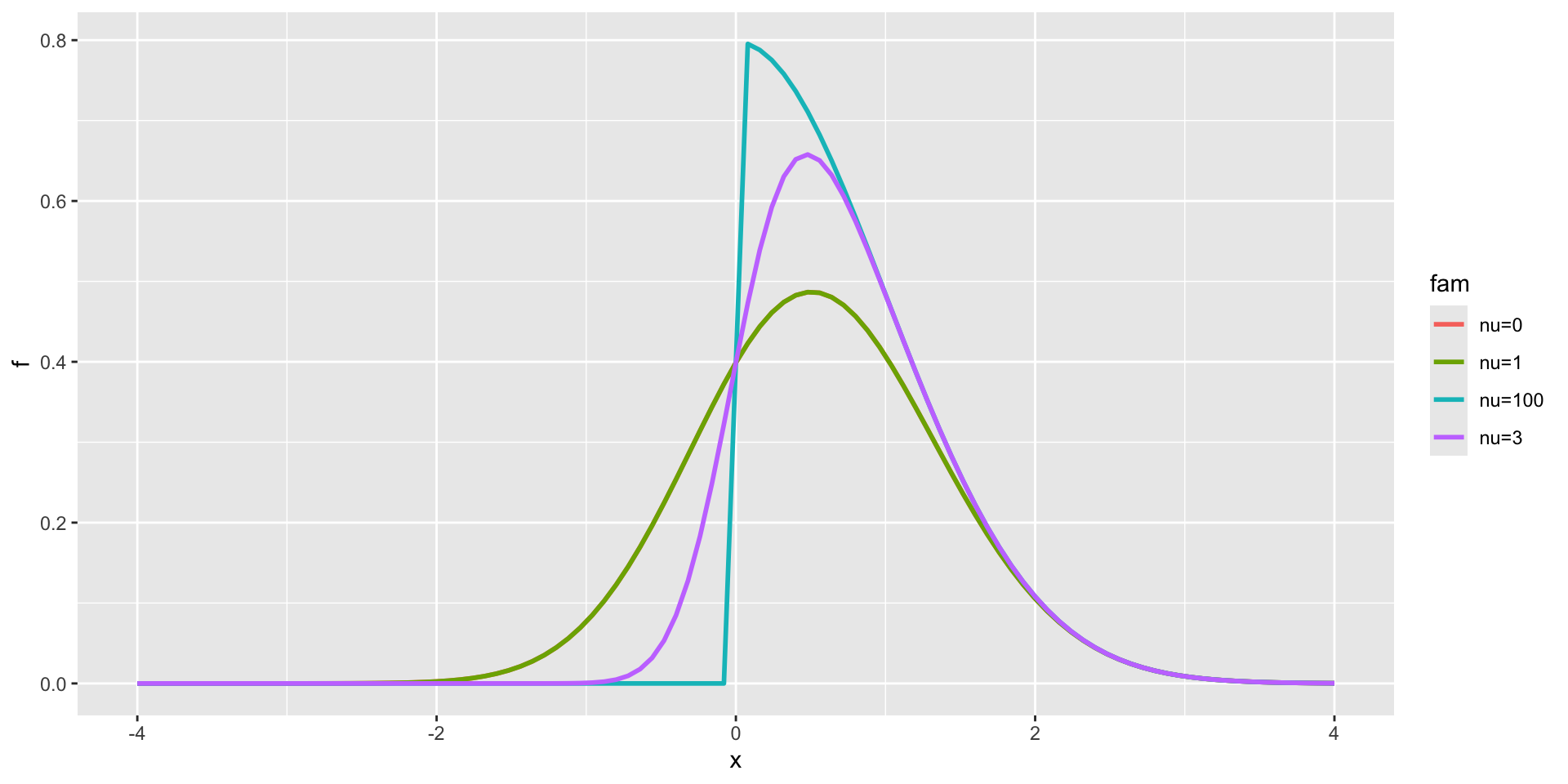
3 parameter in \(\Re\) (TEST 1)
3 parameter in \(\Re\) PE
Power Exponential
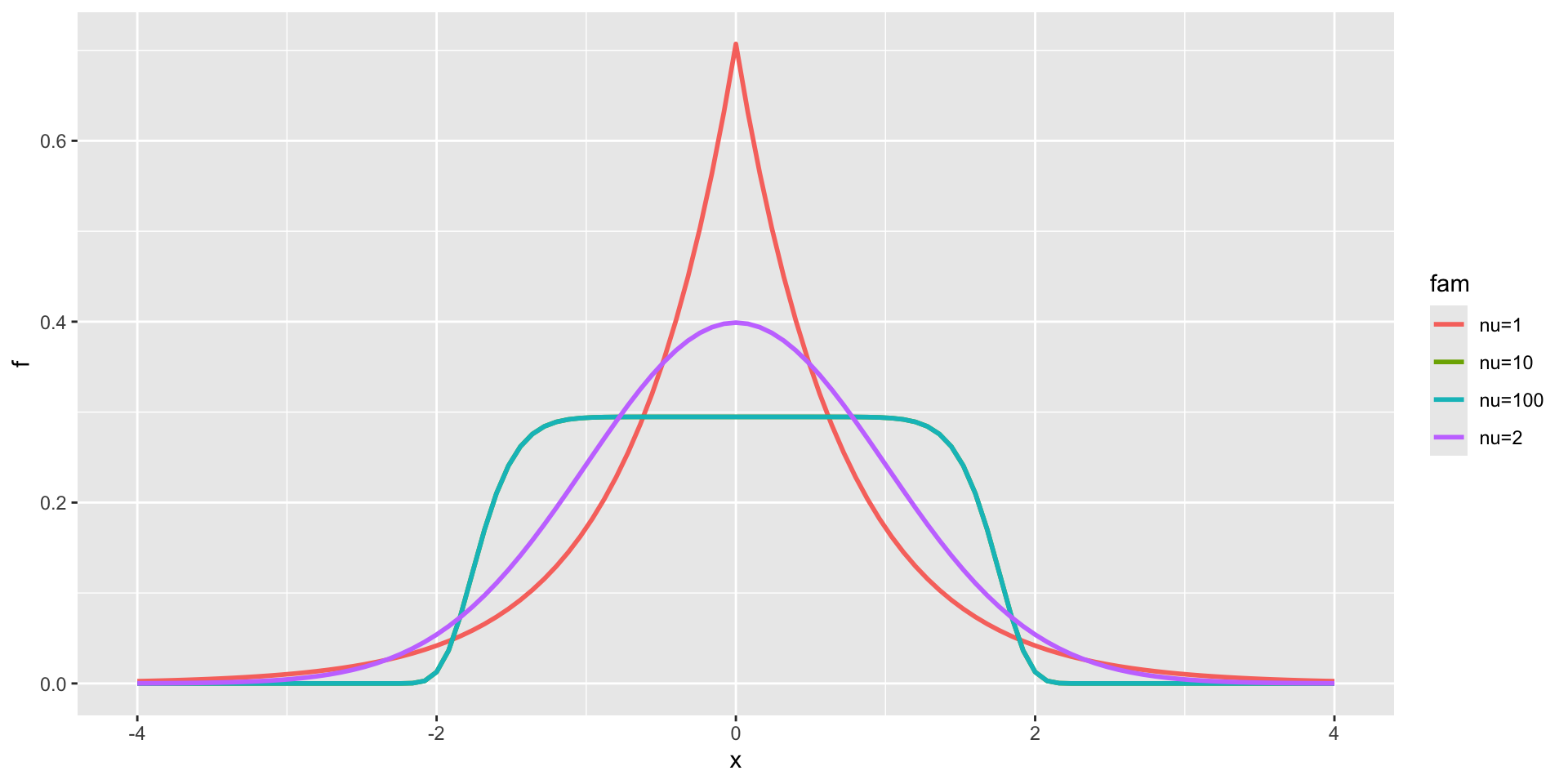
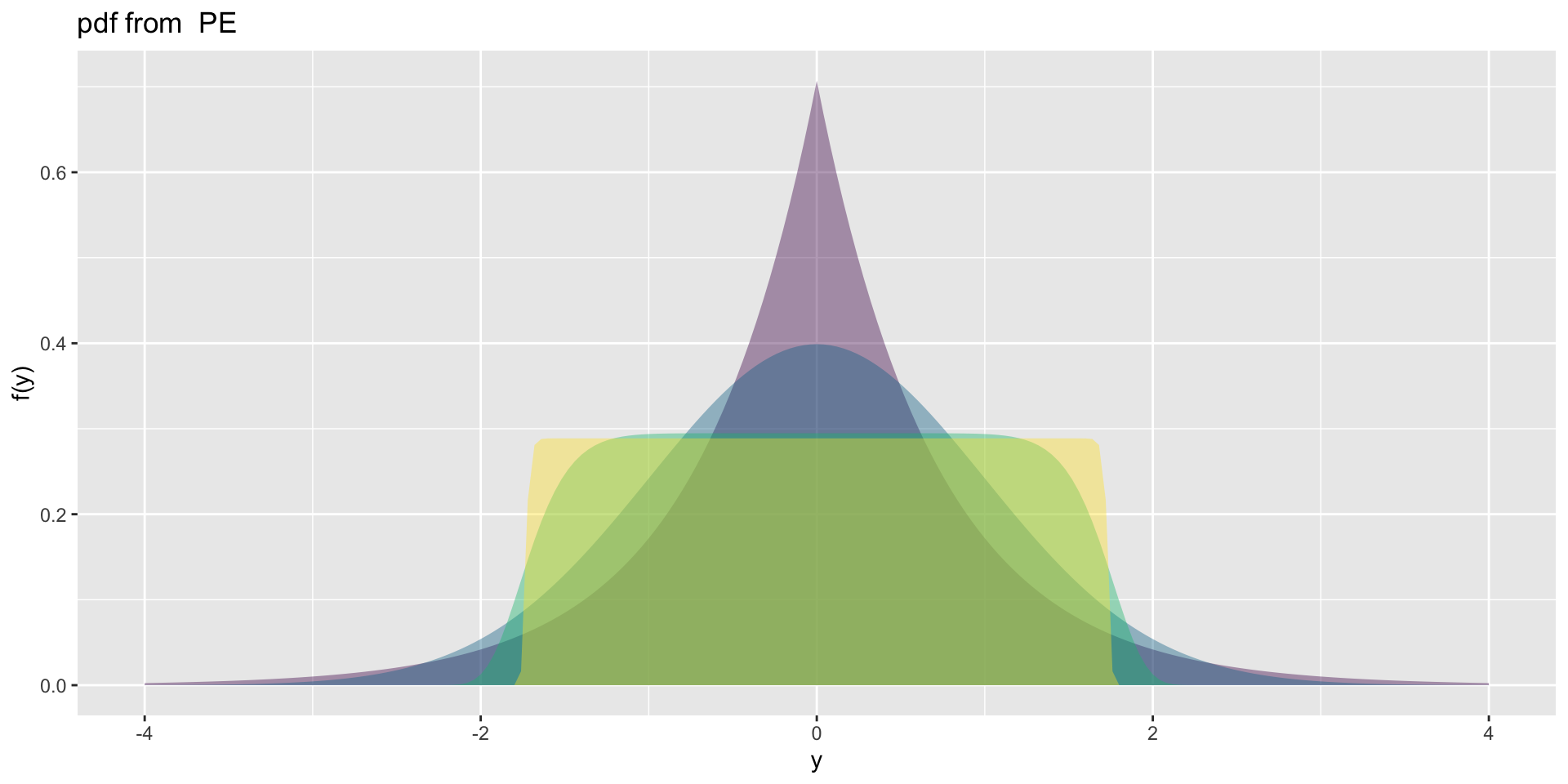
3 parameter in \(\Re\) PE (test)
3 parameter in \(\Re\) TF
t family
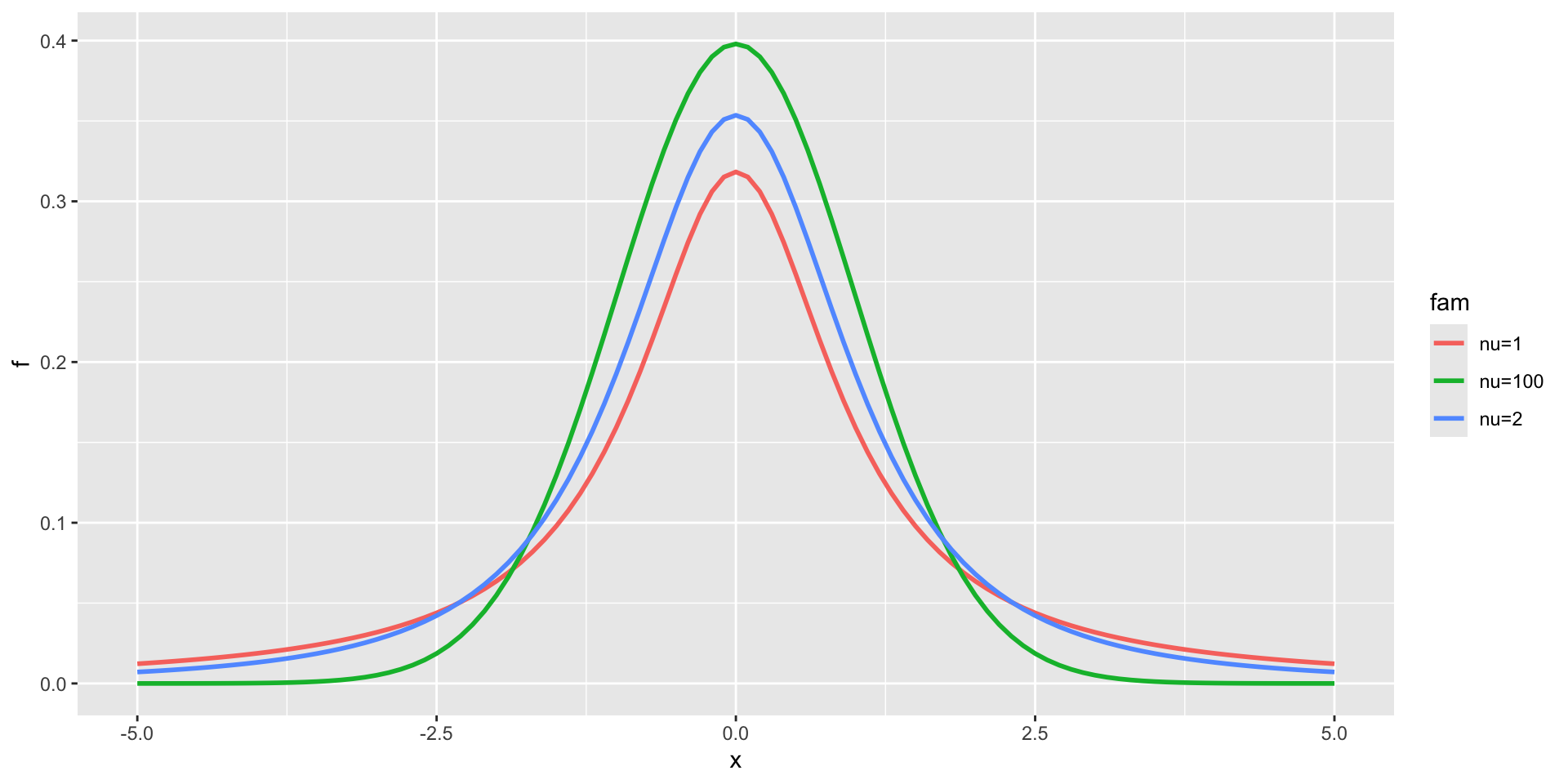
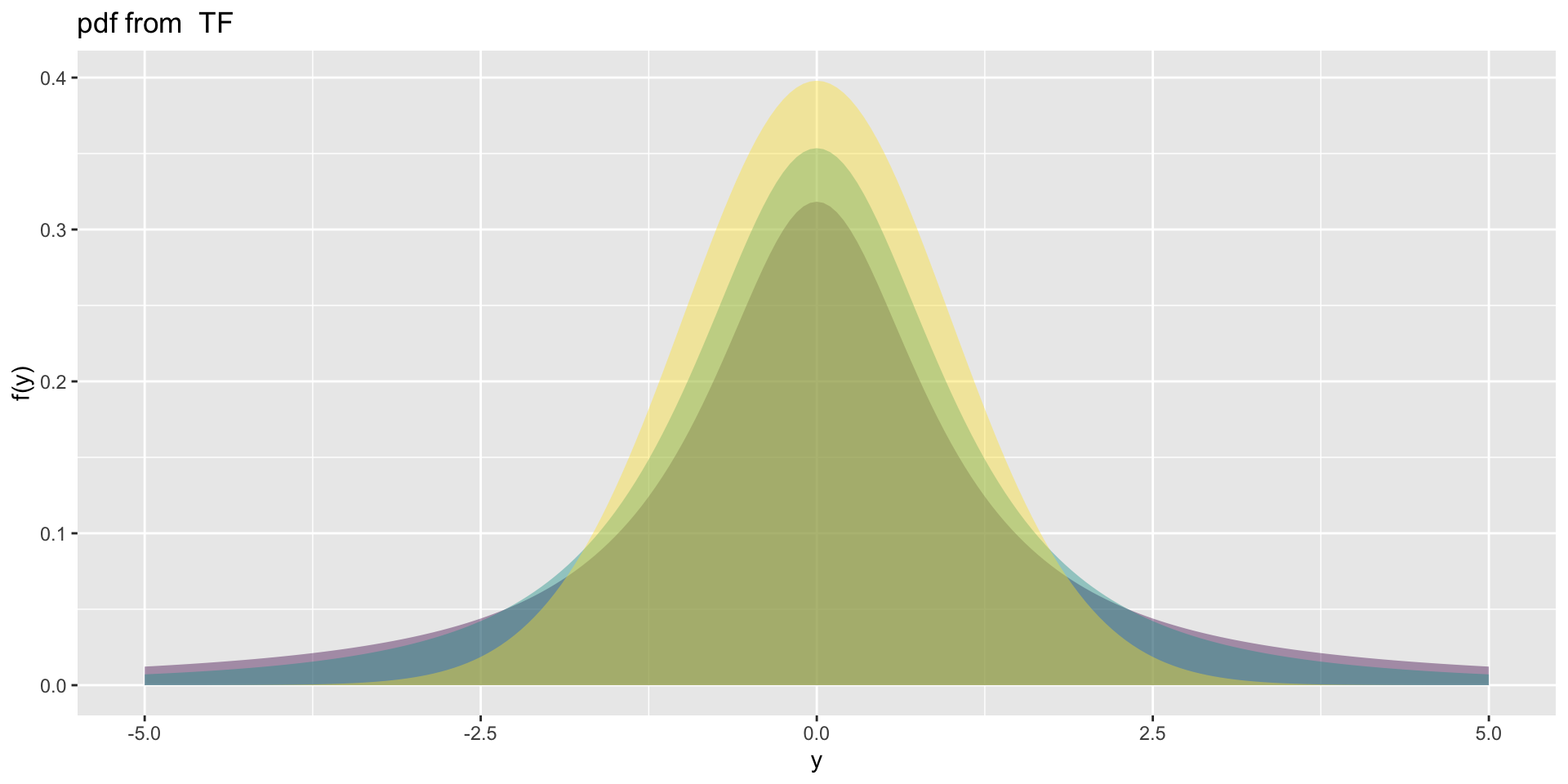
3 parameter in \(\Re\) TF (test)
4 parameter in \(\Re\)
exponential generalised beta type 2,
\(EGB2(\mu,\sigma,\nu,\tau)\) skewness and leptokurtosis;generalised t, GT\((\mu,\sigma, \nu, \tau)\) kurtosis;Johnson's SU, JSU\((\mu,\sigma, \nu, \tau)\) and \(\texttt{JSUo}(\mu,\sigma, \nu, \tau)\) skewness and leptokurtosis;normal-exponential-t\(\texttt{NET}(\mu,\sigma, \nu, \tau)\), robustly location and scaleskew exponential power\(\texttt{SEP1}(\mu,\sigma, \nu, \tau)\), \(\texttt{SEP2}(\mu,\sigma, \nu, \tau)\), \(\texttt{SEP3}(\mu,\sigma, \nu, \tau)\) and \(\texttt{SEP4}(\mu,\sigma, \nu, \tau)\) skewness and lepto-platy;
4 parameter in \(\Re\) (con.)
sinh-arcsinh, \(\texttt{SHASH}(\mu,\sigma, \nu, \tau)\), \(\texttt{SHASHo}(\mu,\sigma, \nu, \tau)\) and \(\texttt{SHASHo2}(\mu,\sigma, \nu, \tau)\) skewness and lepto-platy;skew t, \(\texttt{ST1}(\mu,\sigma, \nu, \tau)\), \(\texttt{ST2}(\mu,\sigma, \nu, \tau)\), \(\texttt{ST3}(\mu,\sigma, \nu, \tau)\), \(\texttt{ST4}(\mu,\sigma, \nu, \tau)\), \(\texttt{ST5}(\mu,\sigma, \nu, \tau)\) and \(\texttt{SST}(\mu,\sigma, \nu, \tau)\) skewness and leptokurtosis.
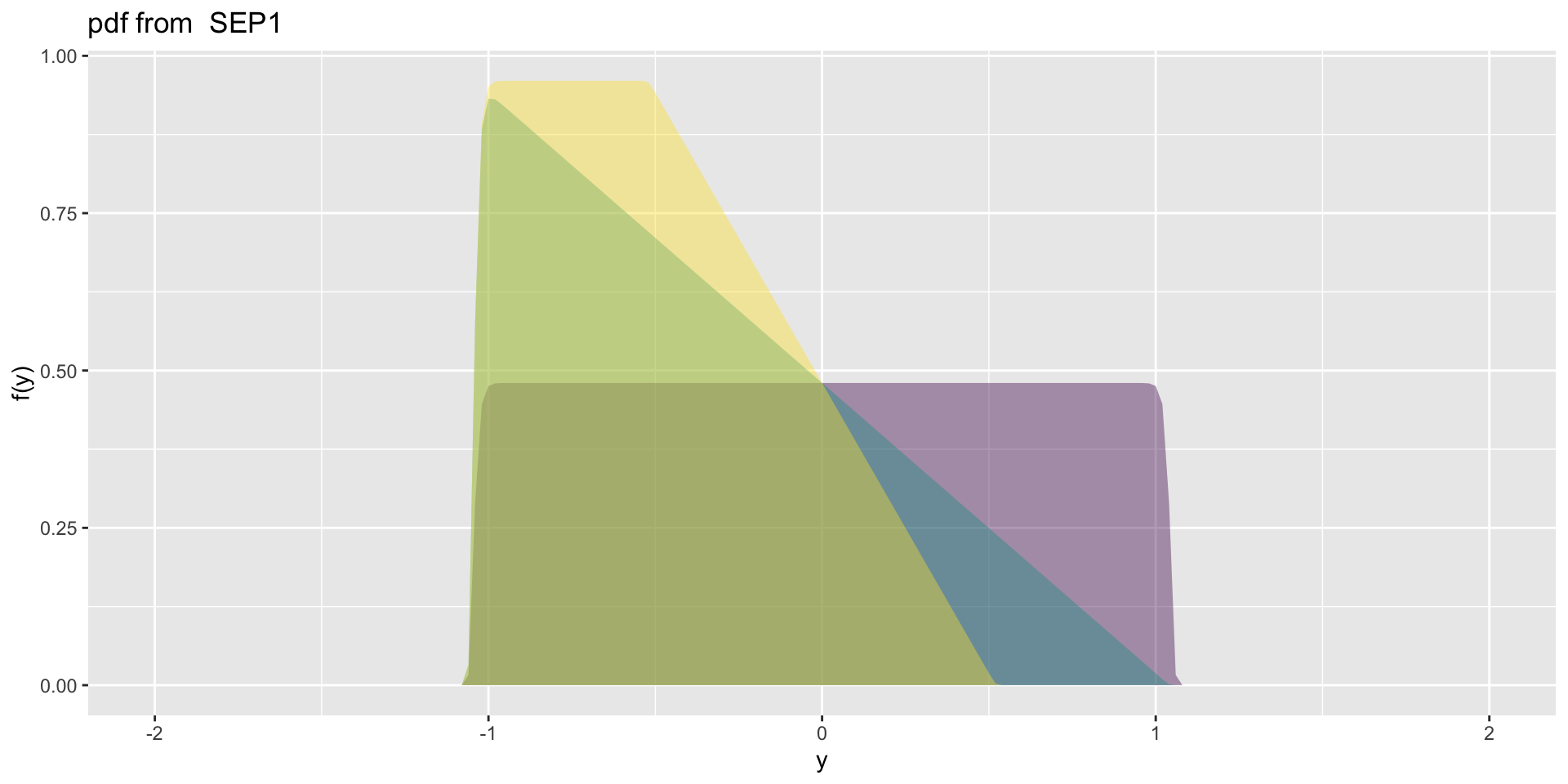
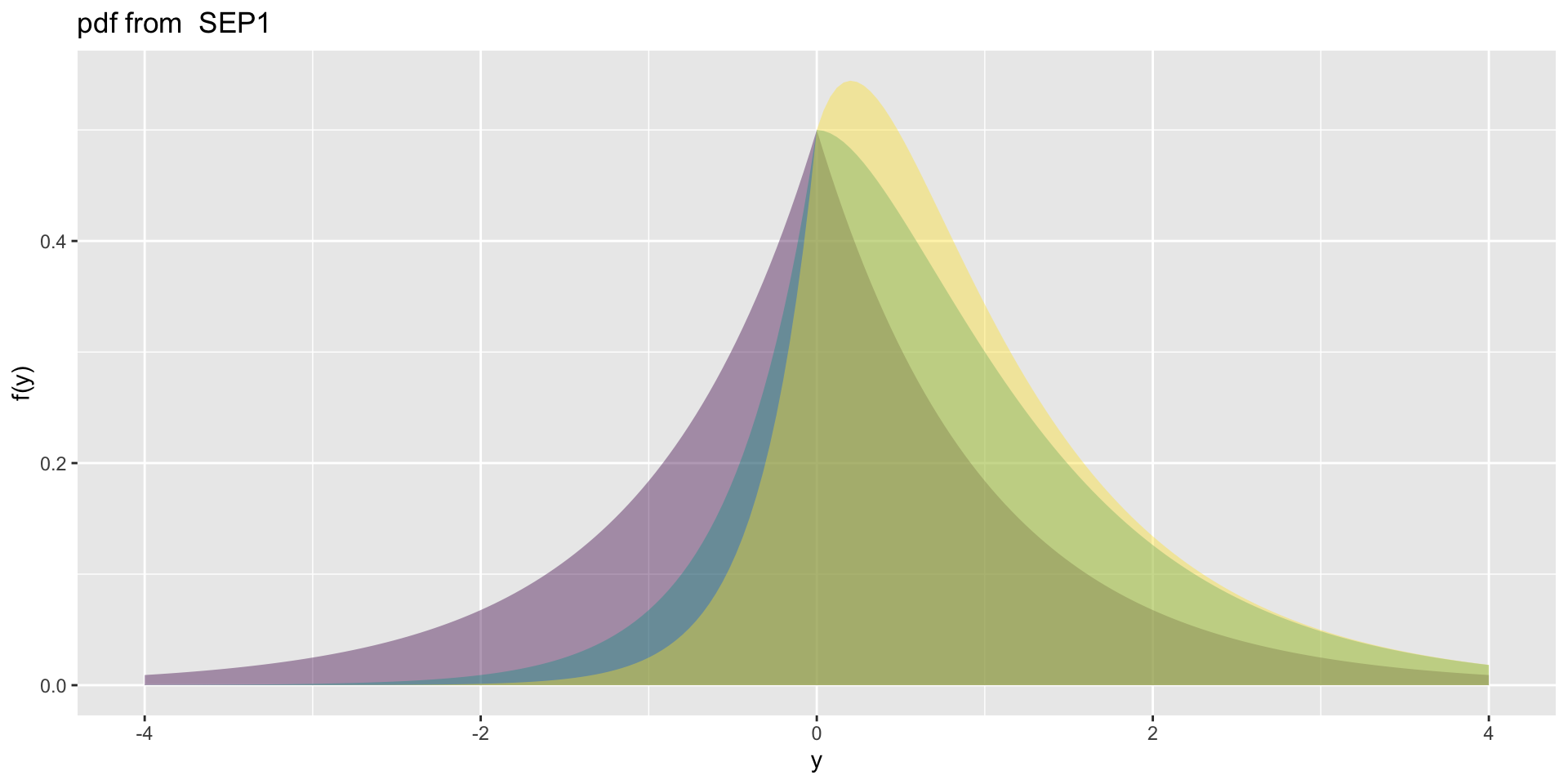
4 parameter in \(\Re\) SEP1
- SEP1\(\mu=0, \sigma=1, \nu=0,1,2, \tau=1\)
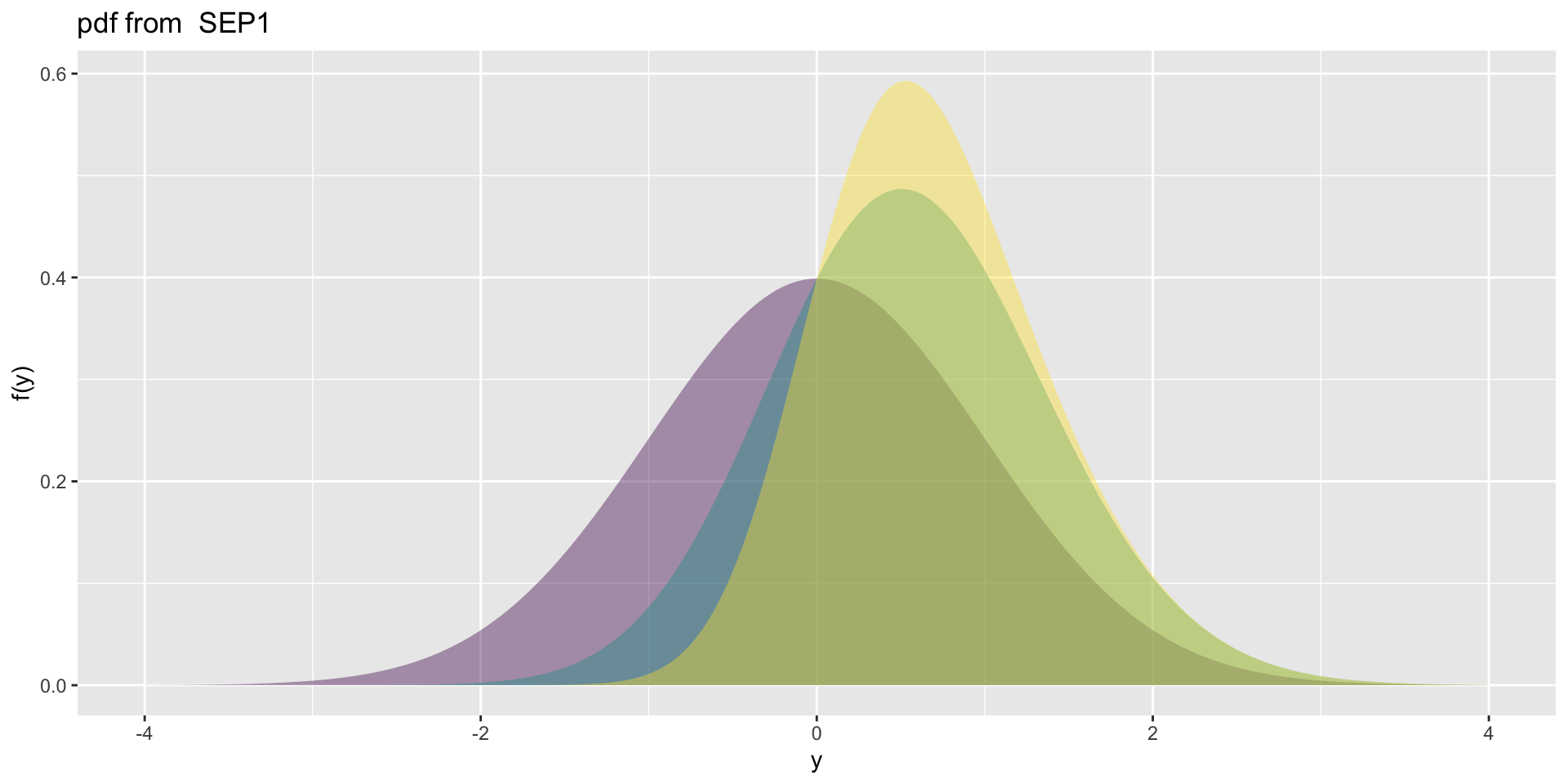
4 parameter in \(\Re\) SEP1
- SEP1\(\mu=0, \sigma=1, \nu=0,1,2, \tau=2\)
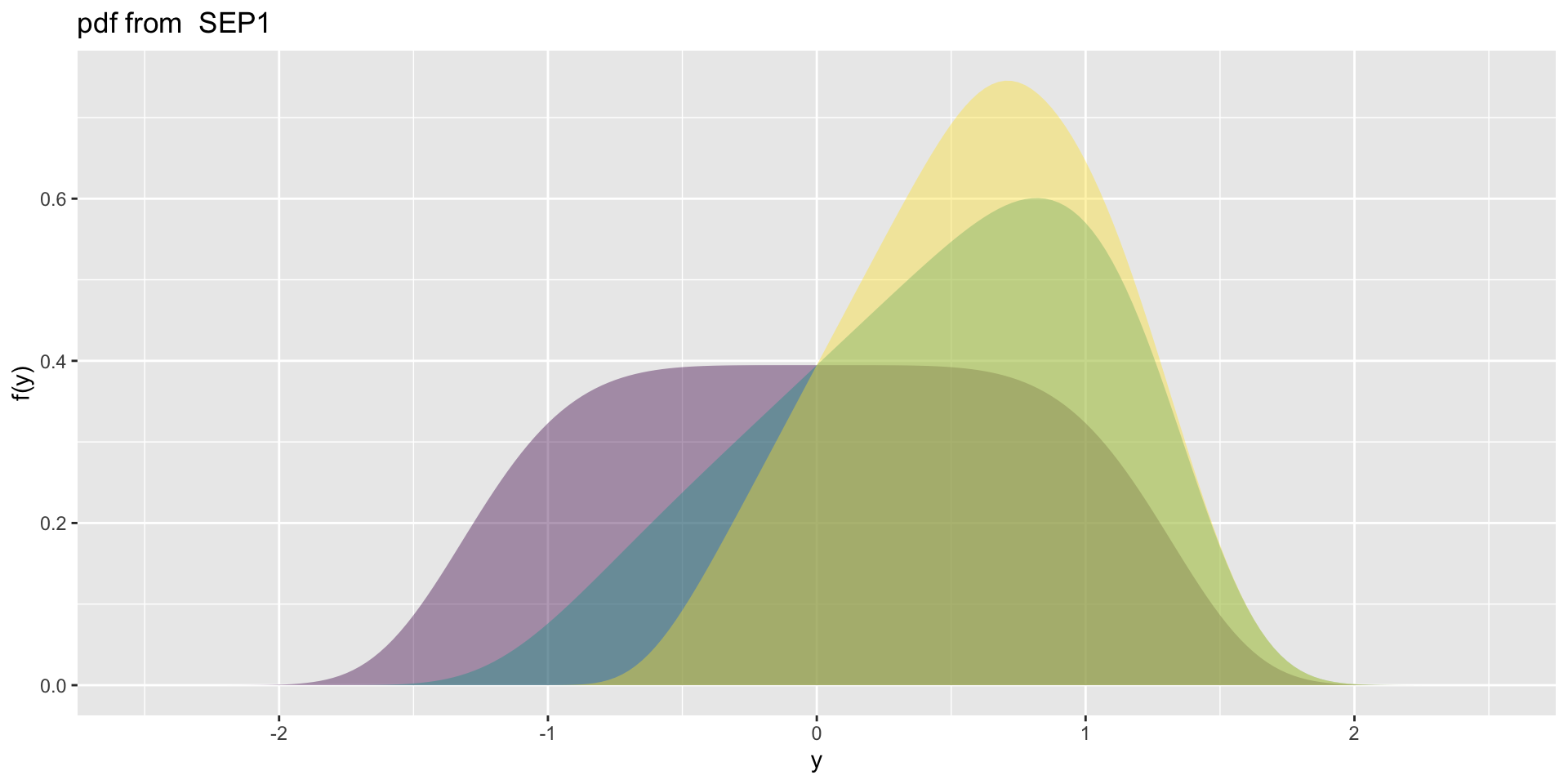
4 parameter in \(\Re\) SEP1
- SEP1\(\mu=0, \sigma=1, \nu=0,1,2, \tau=5\)
4 parameter in \(\Re\) SEP1
4 parameter in \(\Re\) NET
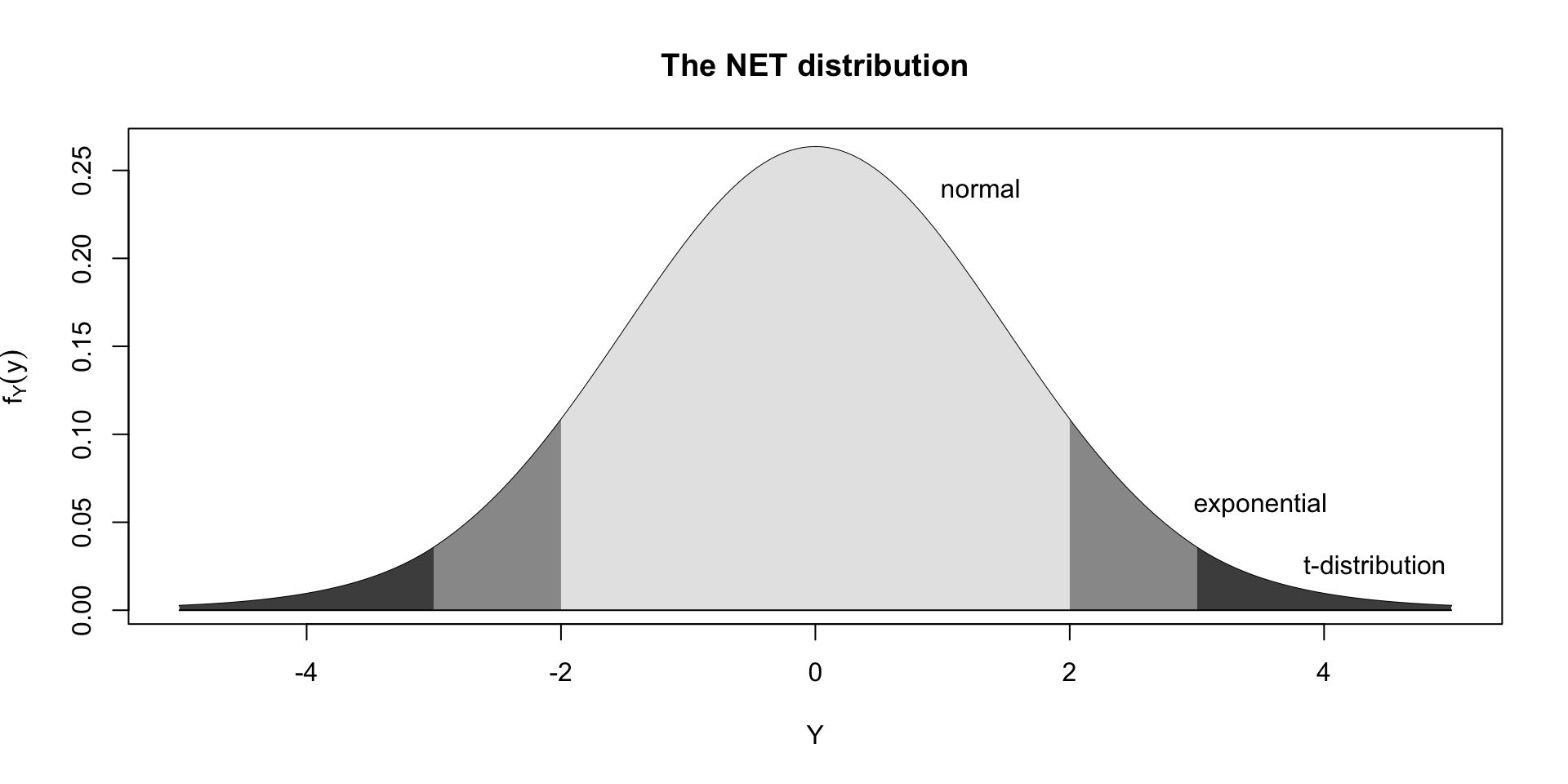
Summary
| Distributions | family | no par. | skewness | kurtosis |
|---|---|---|---|---|
| Exp. Gaussian | exGAUS | 3 | positive | - |
| Exp.G. beta 2 | EGB2 | 4 | both | lepto |
| Gen. t | GT | 4 | (symmetric) | lepto |
| Gumbel | GU | 2 | (negative) | - |
| Johnson’s SU | JSU, JSUo | 4 | both | lepto |
| Logistic | LO | 2 | (symmetric) | (lepto) |
| Normal-Expon.-t | NET | 2,(2) | (symmetric) | lepto |
| Normal | NO-NO2 | 2 | (symmetric) | (meso) |
| Normal Family | NOF | 3 | (symmetric) | (meso) |
| Power Expon. | PE-PE2 | 3 | (symmetric) | both |
| Reverse Gumbel | RG | 2 | positive | |
| Sinh Arcsinh | SHASH, | 4 | both | both |
| Skew Exp. Power | SEP1-SEP4 | 4 | both | both |
| Skew t | ST1-ST5, SST | 4 | both | lepto |
| t Family | TF | 3 | (symmetric) lepto |
Explicit Distributions in positive real line \(\Re^+\)
the scale family
If a random variable is distributed as \[Y\sim D(\mu,\sigma,\nu,\tau)\]
and
\[\varepsilon=(Y/\mu) \sim D(1,\sigma,\nu,\tau)\] then \(Y\) has a scale family
the weibull 3
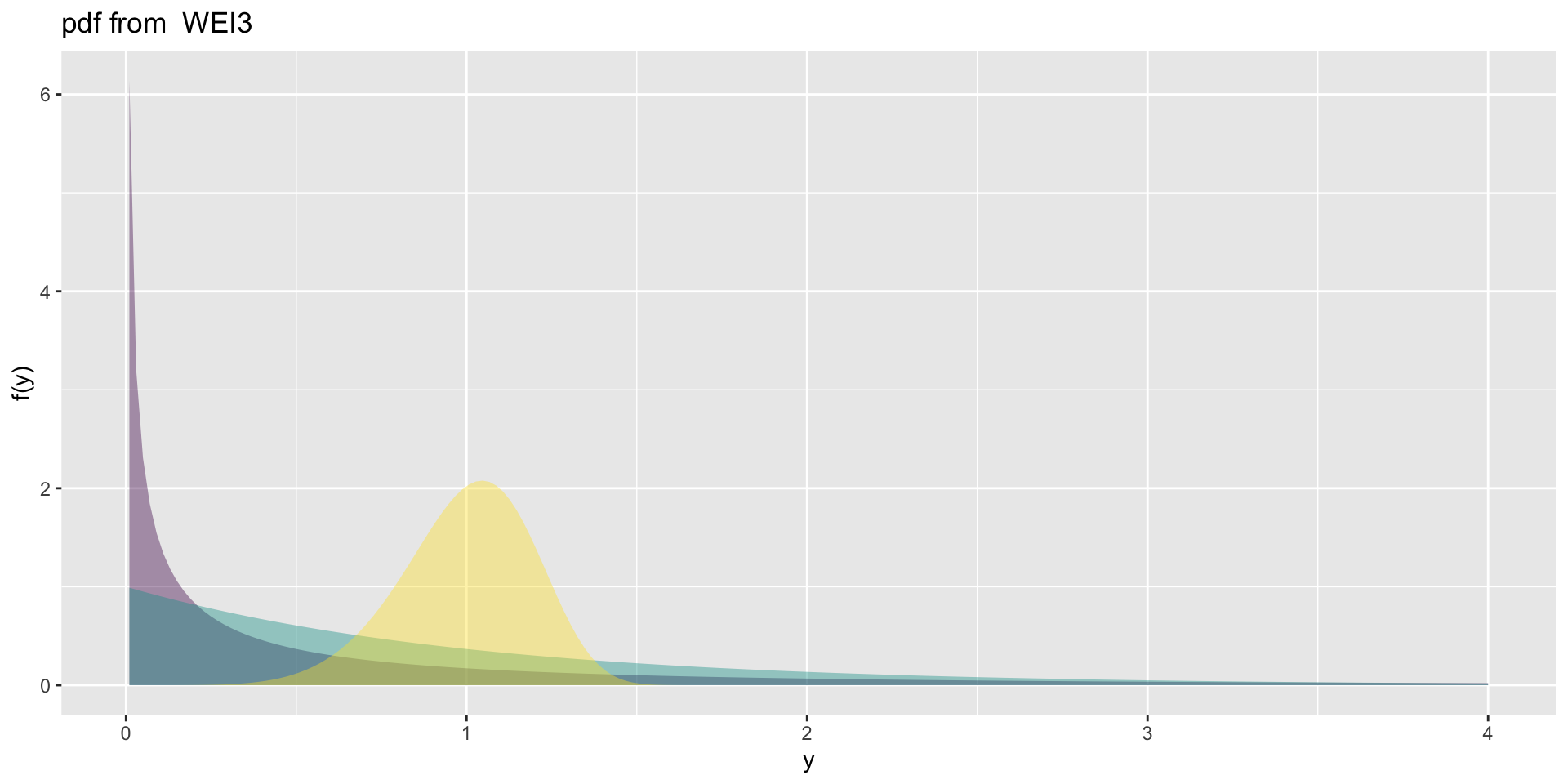
the weibull 3 (con.)
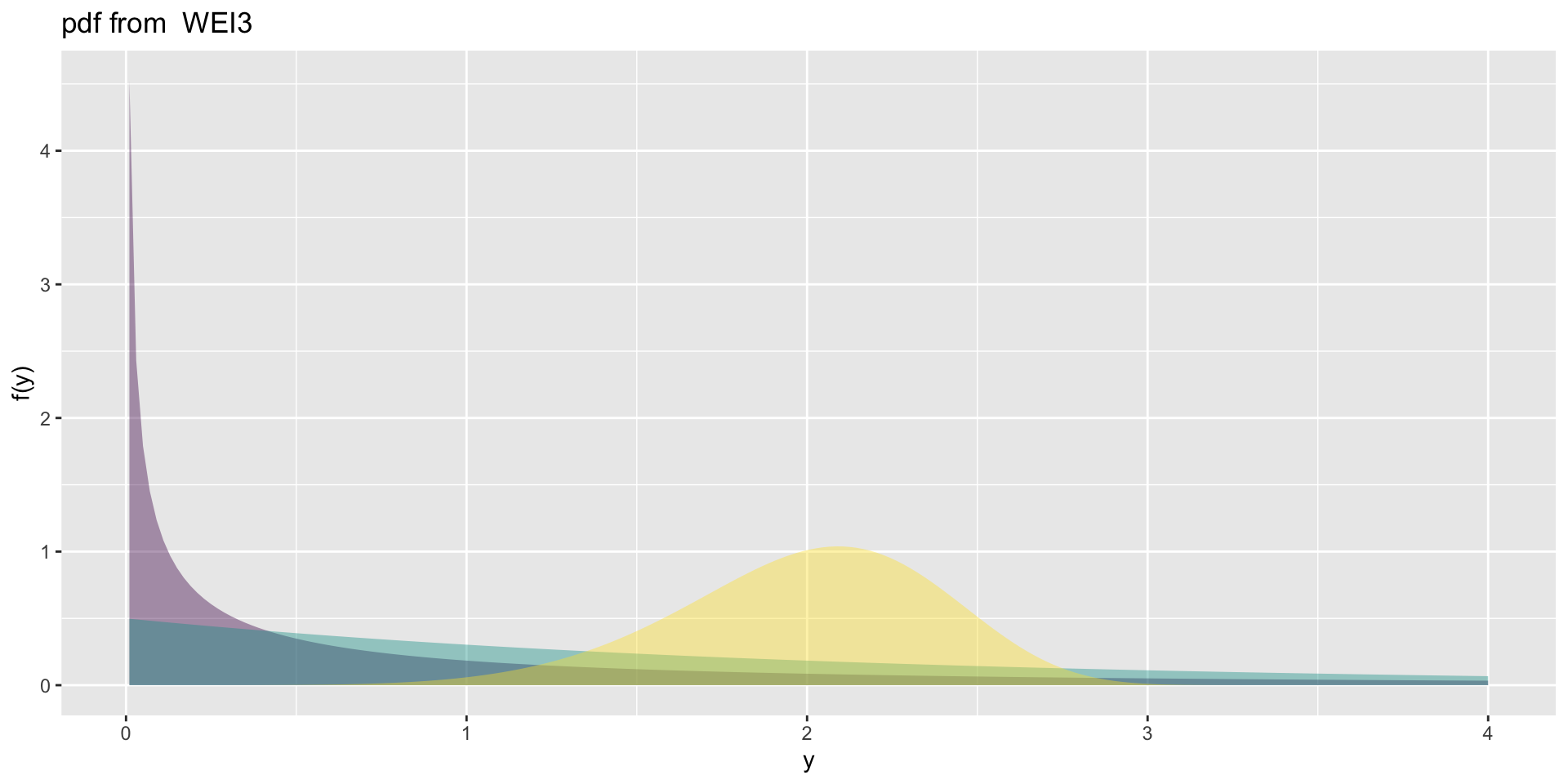
1 and 2 parameter positive real line, \(\Re^+\).
exponencial, \(\texttt{EXP}(\mu,\sigma)\)gamma\(\texttt{GA}(\mu,\sigma)\) member of the exponential family;inverse gamma\(\texttt{IGAMMA}(\mu,\sigma)\);inverse Gaussian, \(\texttt{IG}(\mu,\sigma)\) a member of the exponential family;log-normal\(\texttt{LOGNO}(\mu,\sigma)\) and \(\texttt{LOGNO2}(\mu,\sigma)\).Pareto\(\texttt{PARETO}(\mu,\sigma)\), \(\texttt{PARETO2o}(\mu,\sigma)\) and \(\texttt{GP}(\mu,\sigma)\) for heavy tail;Weibull\(\texttt{WEI}(\mu,\sigma)\), \(\texttt{WEI2}(\mu,\sigma)\) and \(\texttt{WEI3}(\mu,\sigma)\) used in survival analysis.
Weibull survival and hazard
3 parameter positive real line \(\Re^+\)
Box-Cox Cole and Green, \(\texttt{BCCG}(\mu, \sigma, \nu)\) known also as theLMSmethod in centile estimation.generalised gamma, \(\texttt{GG}(\mu, \sigma, \nu)\)generalised inverse Gaussian, \(\texttt{GIG}(\mu, \sigma, \nu)\)log-normal family, \(\texttt{LNO}(\mu, \sigma, \nu)\) based on the standard Box-Cox transformation
the BCCG
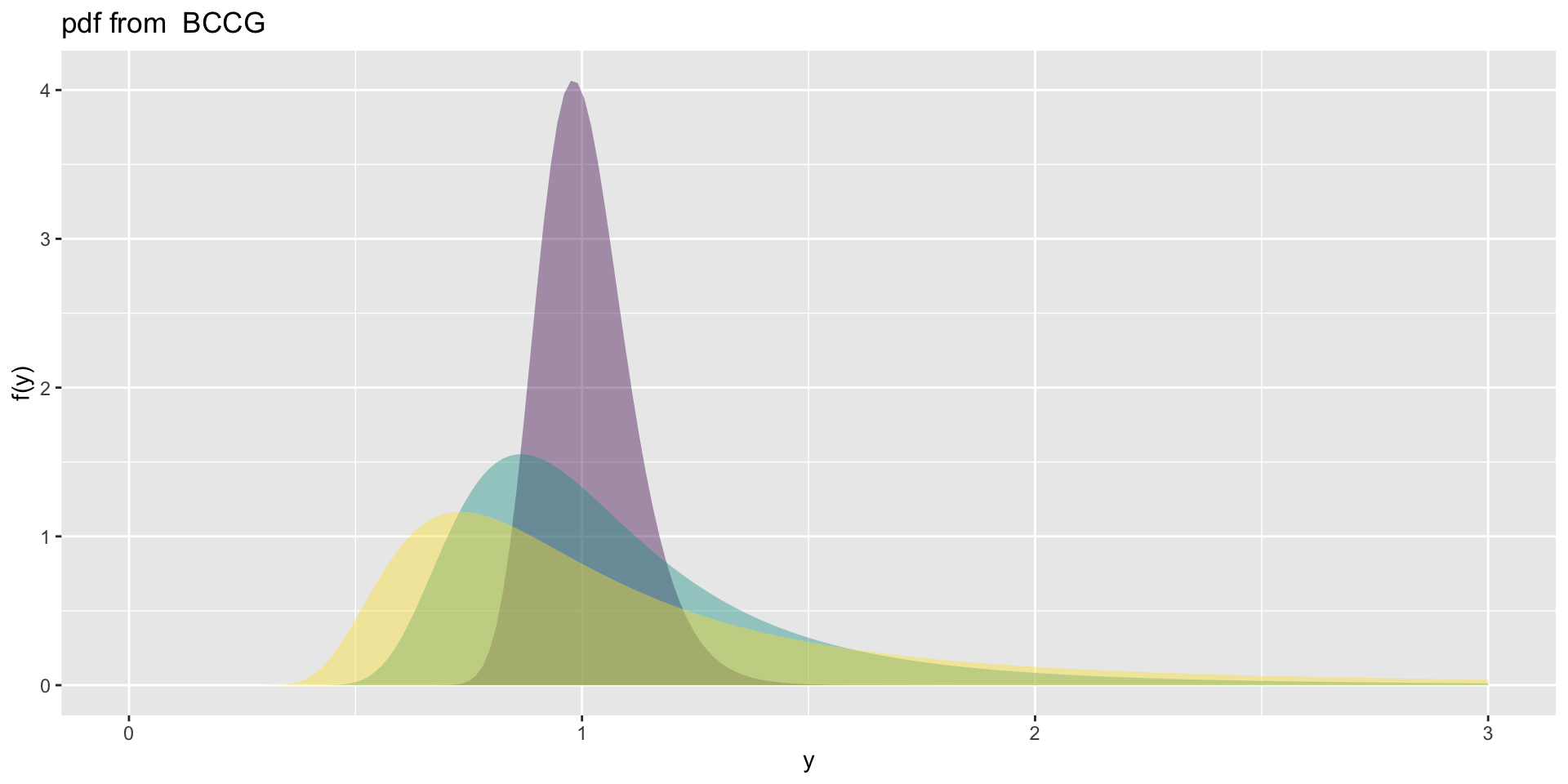
the BCCG (con.)
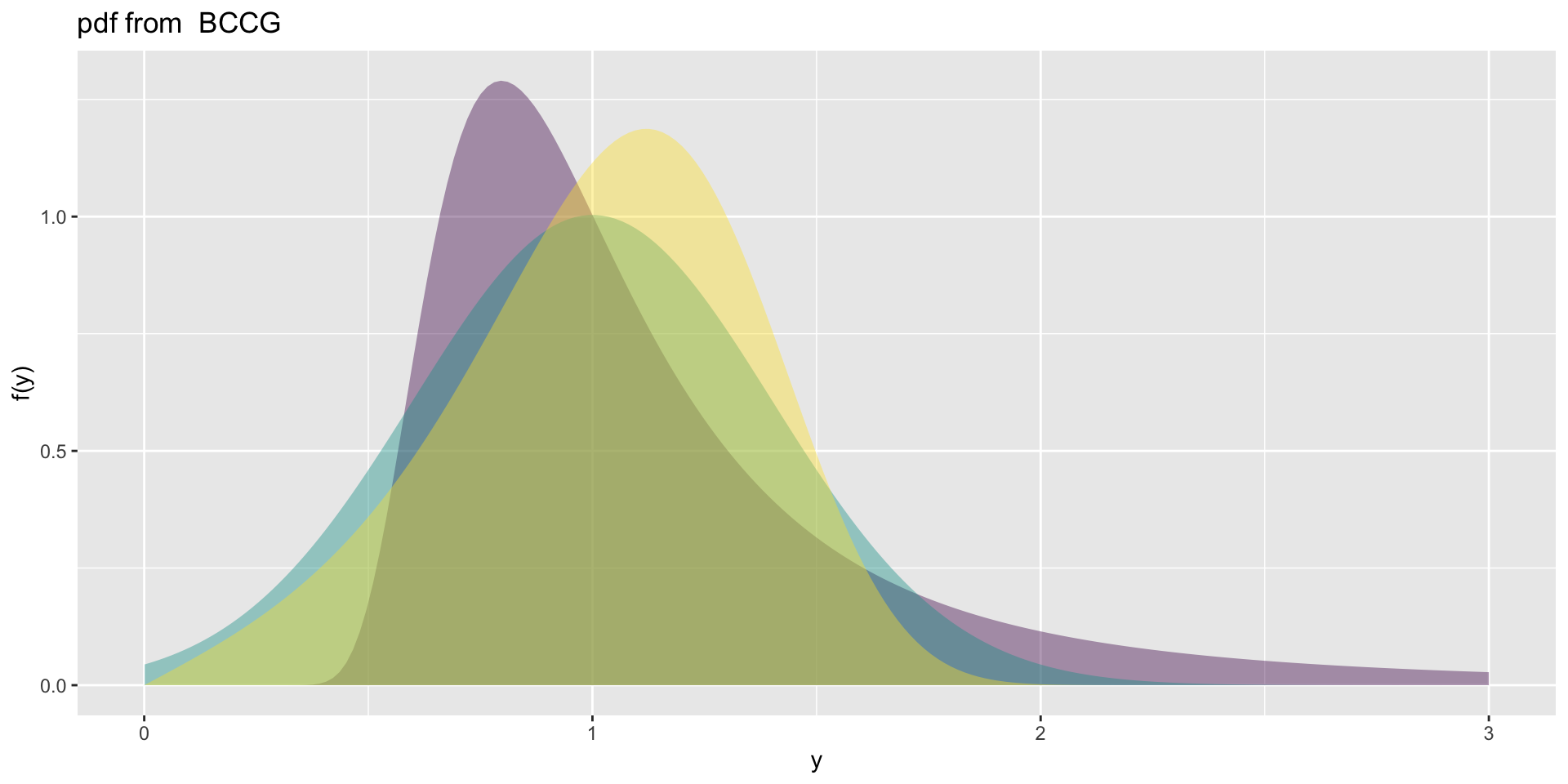
4 parameter positive real line \(\Re^+\)
Box-Cox power exponential, \(\texttt{BCPE}(\mu,\sigma,\nu,\tau)\) and \(\texttt{BCPEo}(\mu,\sigma,\nu,\tau)\) skewness and platy-lepto;Box-Cox t, \(\texttt{BCT}(\mu,\sigma,\nu,\tau)\) and \(\texttt{BCTo}(\mu,\sigma,\nu,\tau)\) skewness and leptokurtosis;generalised beta type 2, \(\texttt{GB2}(\mu,\sigma,\nu,\tau)\) skewness and platy-lepto.
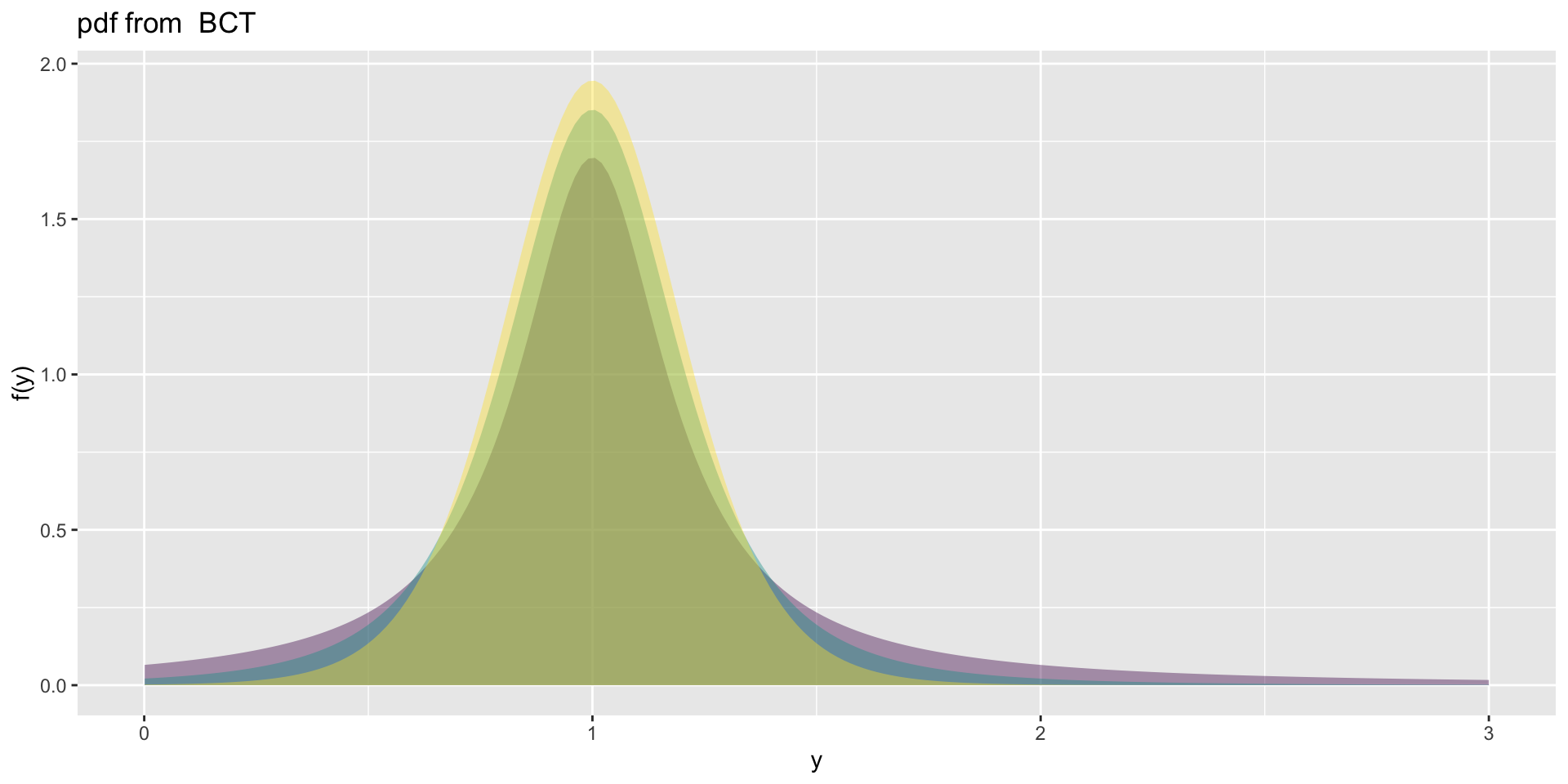
the BCT
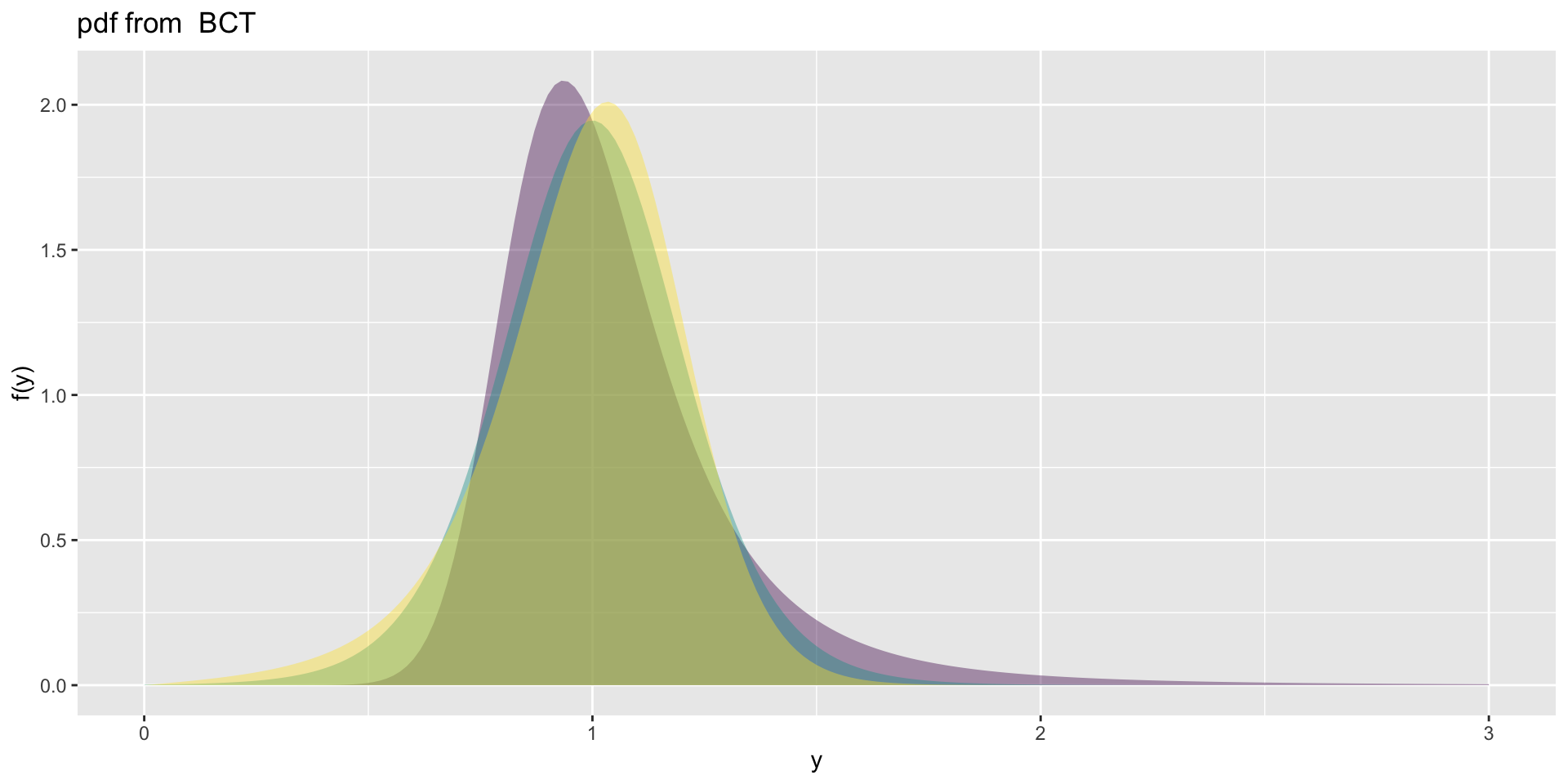
the BCT (con.)
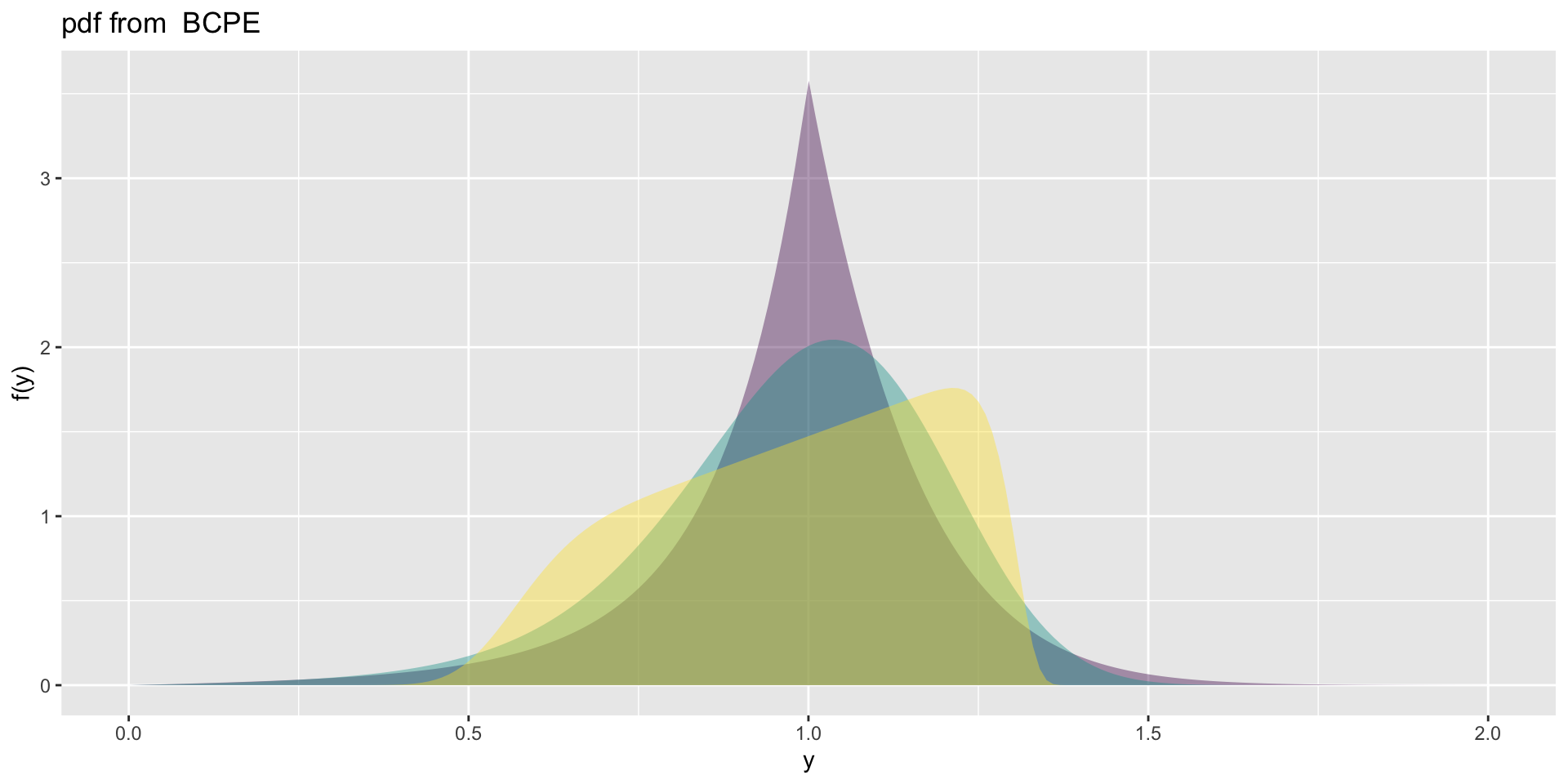
the BCPE
Summary
| Distributions | family | no par. | skewness | kurtosis |
|---|---|---|---|---|
| BCCG | BCCG |
3 | both | |
| BCPE | BCPE |
4 | both | both |
| BCT | BCT |
4 | both | lepto |
| Exponential | EXP |
1 | (positive) | - |
| Gamma | GA |
2 | (positive) | - |
| Gen. Beta type 2 | GB2 |
4 | both | both |
| Gen. Gamma | GG-GG2 |
3 | positive | - |
| Gen. Inv. Gaussian | GIG |
3 | positive | - |
| Inv. Gaussian | IG |
2 | (positive) | - |
| Log Normal | LOGNO |
2 | (positive) | - |
| Log Normal family | LNO |
2,(1) | positive | |
| Reverse Gen. Extreme | RGE |
3 | positive | - |
| Weibull | WEI, WEI2, WEI3 |
2 | (positive) |
{.striped .hover}
Explicit Distributions on \(\Re_{(0,1)}\)
Distributions on \(\Re_{(0,1)}\)
| Distributions | family | no par. | skewness/kurtosis |
|---|---|---|---|
| beta | BE |
2 | (both) |
| beta original | BEo |
2 | (both) |
| generalized beta type 1 | GB1 |
4 | both |
| logit normal | LOGITNO |
2 | (both) |
| simplex | SIMPEX |
2 | (both) |
the BE
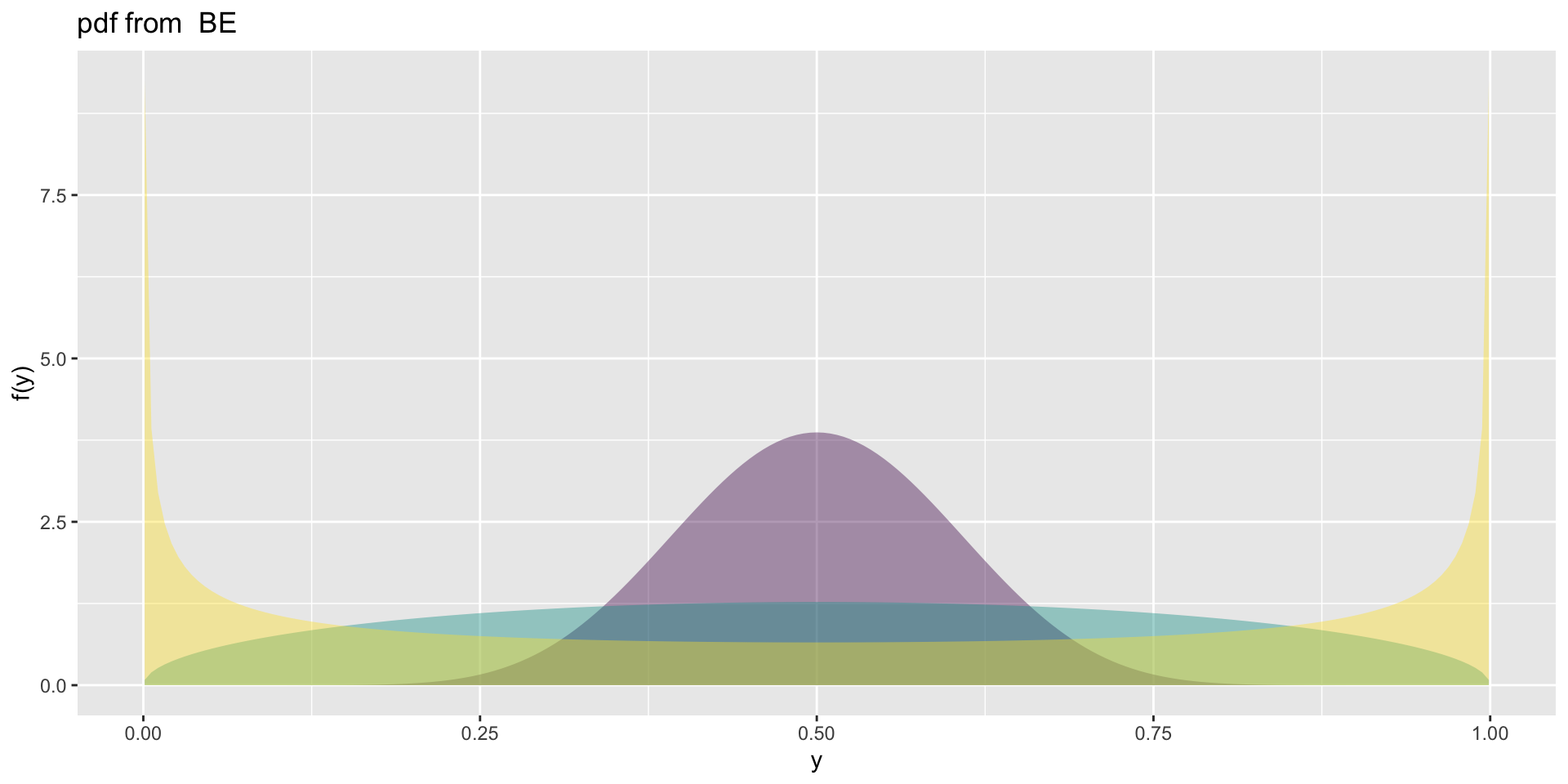
Choosing distributions
Choosing distribution
global deviance
\[ \begin{split} GDEV=& -2\ell(\boldsymbol{\hat{\theta}}) =& -2 \sum_{i=1}^n \log f(y_i |\boldsymbol{\hat{\theta}}) \end{split}\]
generalized Akaike information criterion \[
GAIC(k) = GDEV + k \cdot df
\] special cases \[\begin{split}
{\rm AIC} &= \texttt{GDEV}+2 \, df\\
{\rm SBC} &= \texttt{GDEV}+ \log n \cdot df\ .
\end{split}\]
Choosing distribution
prediction global deviance
\({\widetilde{ y}}\) indicates the response variable values at the validation (or test) sample,
How to fit distributions in R
optim()ormle()R functions requiring initial parameter valuesgamlsssML()fits a distribution (using MLE) on the response with no explanatory variablesgamlss()fits a distribution on the response using RS or CG or mixed algorithms,histDist()fits a distribution usinggamlsssML(), and plots a histogram of the response with the fitted distributionfitDist()fits a set of distributions to the response and chooses the one with the smallest GAICchooseDist()fits a set of distributions on a fitted model and chooses the one with the smallest GAIC
Example: DAX returns data
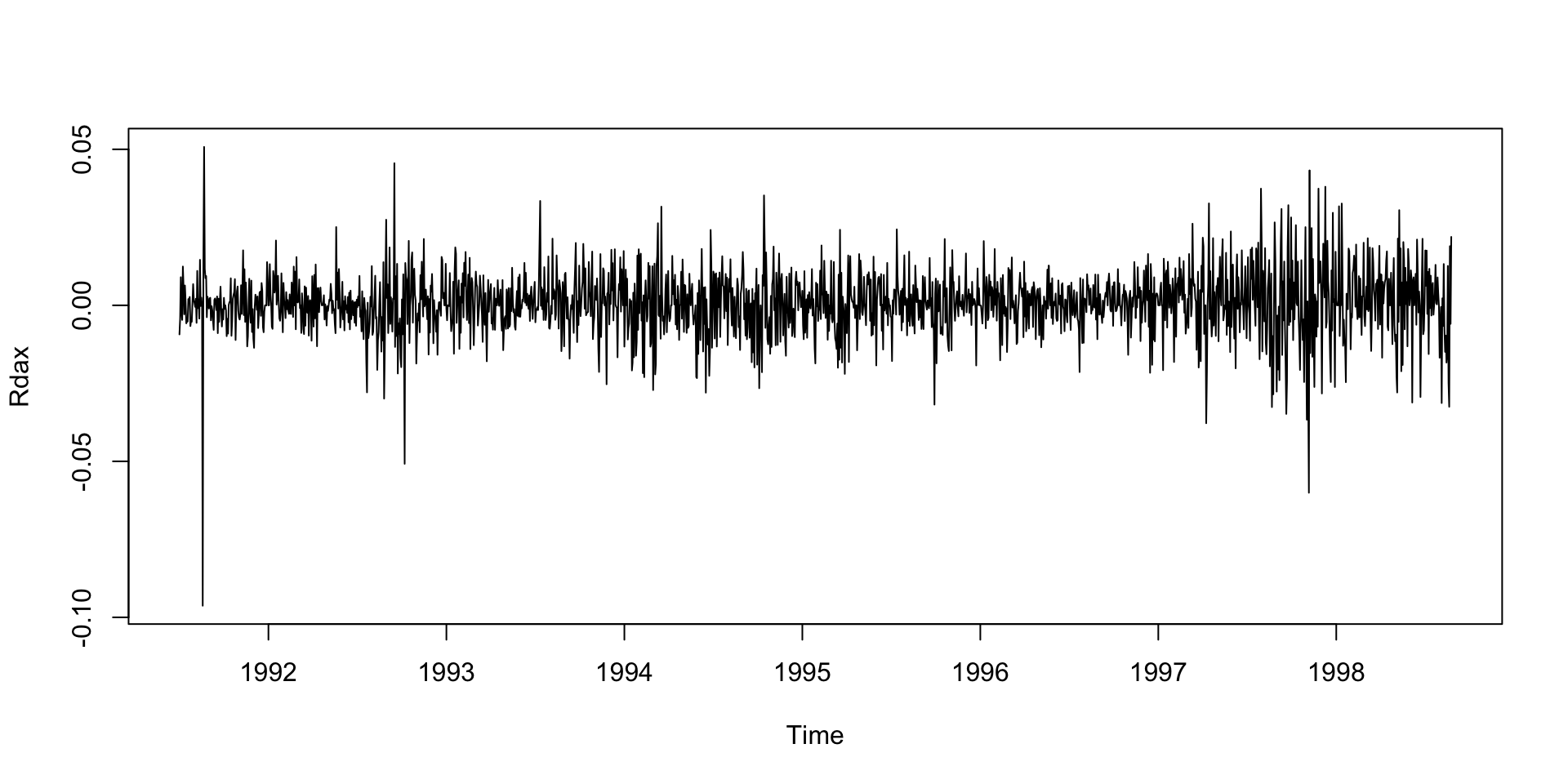
Example: DAX returns data (con.)
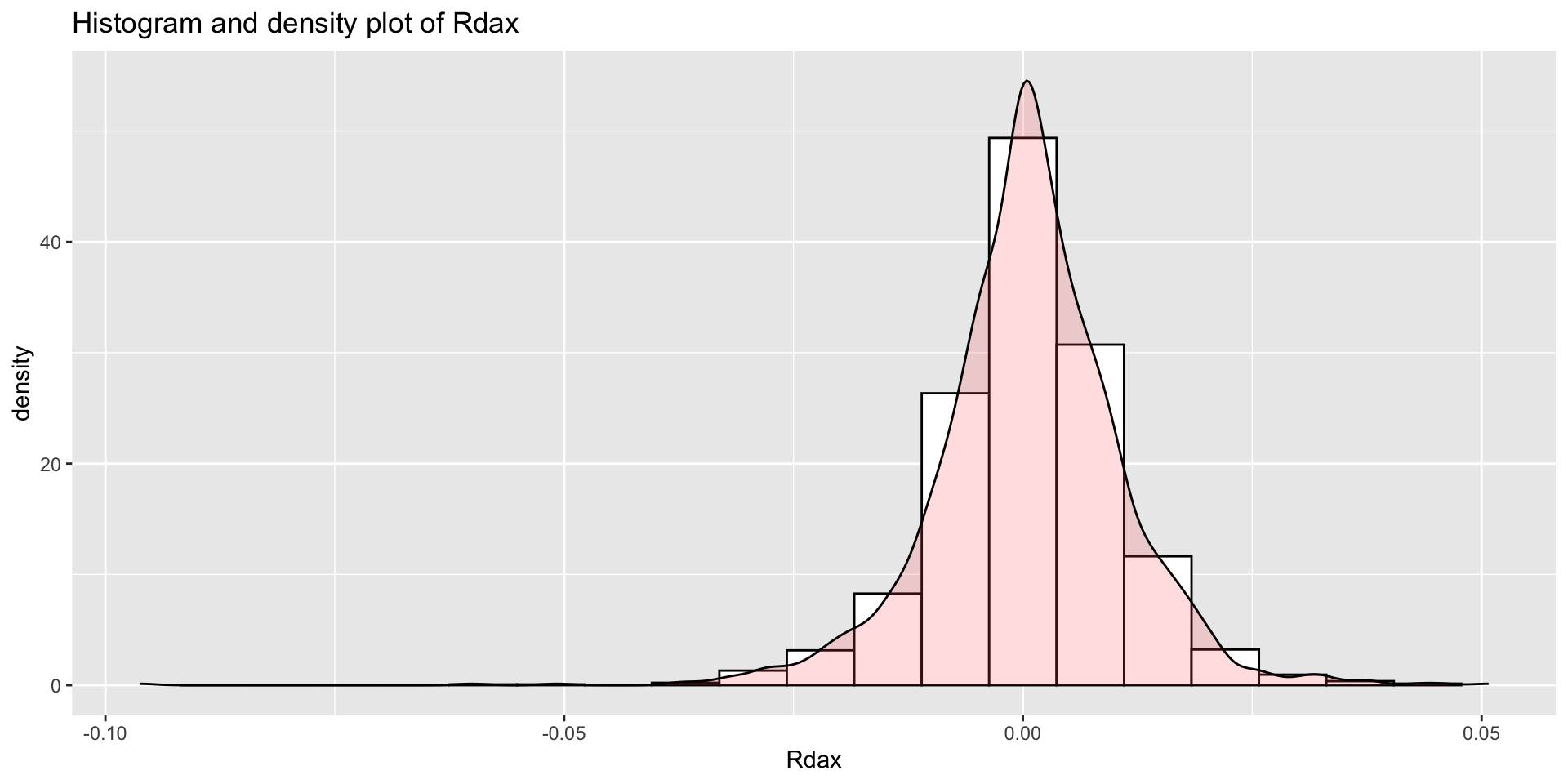
fitDist()
GT SEP2 PE PE2 JSU JSUo SEP1
-11967.683 -11965.533 -11962.464 -11962.464 -11961.477 -11961.477 -11961.305
SEP4 SEP3 TF2 TF ST4 ST1 EGB2
-11961.274 -11960.823 -11960.644 -11960.644 -11960.153 -11959.833 -11959.701
ST5 ST2 ST3 SST SHASH SHASHo2 SHASHo
-11959.479 -11959.280 -11958.866 -11958.866 -11957.538 -11956.241 -11956.241
LO NET SN1 SN2 NO exGAUS GU
-11932.112 -11919.175 -11759.871 -11739.127 -11733.208 -11733.171 -11262.120
RG GA GG GB2 BCTo BCCGo BCPEo
-10249.878 -7506.758 -7506.702 -7506.330 -7505.499 -7503.496 -7503.111
EXP PARETO2 PARETO2o IG IGAMMA
-7481.488 -7479.488 -7479.487 -6705.703 -6486.337 chooseDist()
GAMLSS-RS iteration 1: Global Deviance = -11737.208 eps = 0.000000 minimum GAIC(k= 2 ) family: GT
minimum GAIC(k= 3.84 ) family: GT
minimum GAIC(k= 7.53 ) family: PE GAIG with k= 2 GT SEP2 PE PE2 JSU SEP1
-11966.32 -11965.46 -11962.46 -11962.21 -11961.45 -11961.16 chooseDist()(con.)
[1] 0.0007286232[1] 0.01005041[1] 3.633405[1] 1.603723histDist()
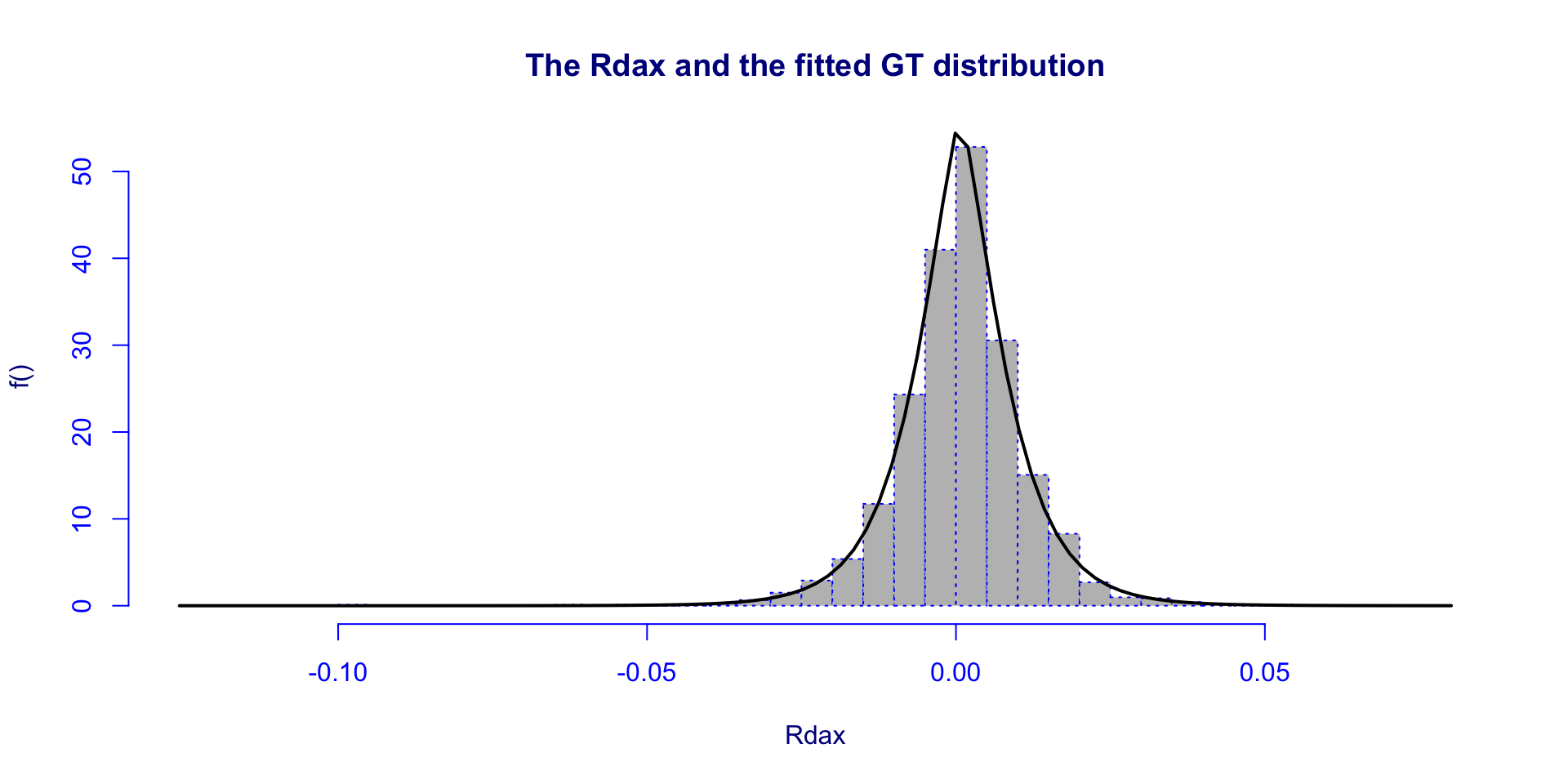
practical 2
END


 The Books
The Books

www.gamlss.com